Open methodology for “Exact approach for convex adjustable robust optimization > Resource Allocation Problem (RAP)
Loading the data
The raw results can be found in the file results.csv
with the following columns:
- tag: a tag which should always equal “result” used to
grepthe result line in the execution log file. - instance: the name or path of the instance.
- n_servers: the number of servers in the instance.
- n_clients: the number of clients in the instance.
- Gamma: the value of \(\Gamma\) which was used.
- deviation: the maximum deviation, in percentage, from the nominal demand.
- time_limit: the time limit which was used when solving the instance.
- method: the name of the method which was used to solve the instance.
- total_time: the total time used to solve the instance.
- master_time: the time spent solving the master problem.
- separation_time: the time spent solving the separation problem.
- best_bound: the best bound found.
- iteration_count: the number of iterations.
- fail: should be empty if the execution of the algorithm went well.
We start by reading the file and by removing the “tag” column.
results = read.table("./new-results.csv", header = FALSE, sep = ',')
colnames(results) <- c("tag", "instance", "n_servers", "n_clients", "Gamma", "deviation", "time_limit", "method", "use_heuristic", "total_time", "master_time", "separation_time", "best_bound", "iteration_count", "fail")
results$tag = NULLThen, we compute the percentage which \(\Gamma\) represents for the total number of clients.
results$p = results$Gamma / results$n_clients
results$p = ceiling(results$p / .05) * .05results$size = paste0("(", results$n_servers, ",", results$n_clients, "), ", results$p)results$method_extended = paste0(results$method, ", ", results$use_heuristic)We also get rid of instances which were too large to be solved by both approaches.
results = results[!(results$n_clients == 40),]
results = results[!(results$n_servers == 25 & results$n_clients == 25),]We then check that all instances were solved without issue by checking the “fail” column.
sum(results$fail)## [1] 62We add a tag for unsolved instances.
time_limit = 7200
if (sum(results$fail) > 0) {
results[results$fail == TRUE,]$total_time = time_limit
}
results$unsolved = results$total_time >= time_limit | results$failAll in all, our result data reads.
Performance profiles and ECDF
We now introduce a function which plots the performance profile of our solvers over a given test set.
add_performance_ratio = function(dataset,
criterion_column = "total_time",
unsolved_column = "unsolved",
instance_column = "instance",
solver_column = "solver",
output_column = "performance_ratio") {
# Compute best score for each instance
best = dataset %>%
group_by(!!sym(instance_column)) %>%
mutate(best_solver = min(!!sym(criterion_column)))
# Compute performance ratio for each instance and solver
result = best %>%
group_by(!!sym(instance_column), !!sym(solver_column)) %>%
mutate(!!sym(output_column) := !!sym(criterion_column) / best_solver) %>%
ungroup()
if (sum(result[,unsolved_column]) > 0) {
result[result[,unsolved_column] == TRUE,output_column] = max(result[,output_column])
}
return (result)
}
plot_performance_profile = function(dataset,
criterion_column,
unsolved_column = "unsolved",
instance_column = "instance",
solver_column = "solver"
) {
dataset_with_performance_ratios = add_performance_ratio(dataset,
criterion_column = criterion_column,
instance_column = instance_column,
solver_column = solver_column,
unsolved_column = unsolved_column)
solved_dataset_with_performance_ratios = dataset_with_performance_ratios[!dataset_with_performance_ratios[,unsolved_column],]
compute_performance_profile_point = function(method, data) {
performance_ratios = solved_dataset_with_performance_ratios[solved_dataset_with_performance_ratios[,solver_column] == method,]$performance_ratio
unscaled_performance_profile_point = ecdf(performance_ratios)(data)
n_instances = sum(dataset[,solver_column] == method)
n_solved_instances = sum(dataset[,solver_column] == method & !dataset[,unsolved_column])
return( unscaled_performance_profile_point * n_solved_instances / n_instances )
}
perf = solved_dataset_with_performance_ratios %>%
group_by(!!sym(solver_column)) %>%
mutate(performance_profile_point = compute_performance_profile_point(unique(!!sym(solver_column)), performance_ratio))
result = ggplot(data = perf, aes(x = performance_ratio, y = performance_profile_point, color = !!sym(solver_column))) +
geom_line()
return (result)
}performance_profile = plot_performance_profile(results, criterion_column = "total_time", solver_column = "method_extended") +
labs(x = "Performance ratio", y = "% of instances") +
scale_y_continuous(limits = c(0, 1)) +
theme_minimal()
print(performance_profile)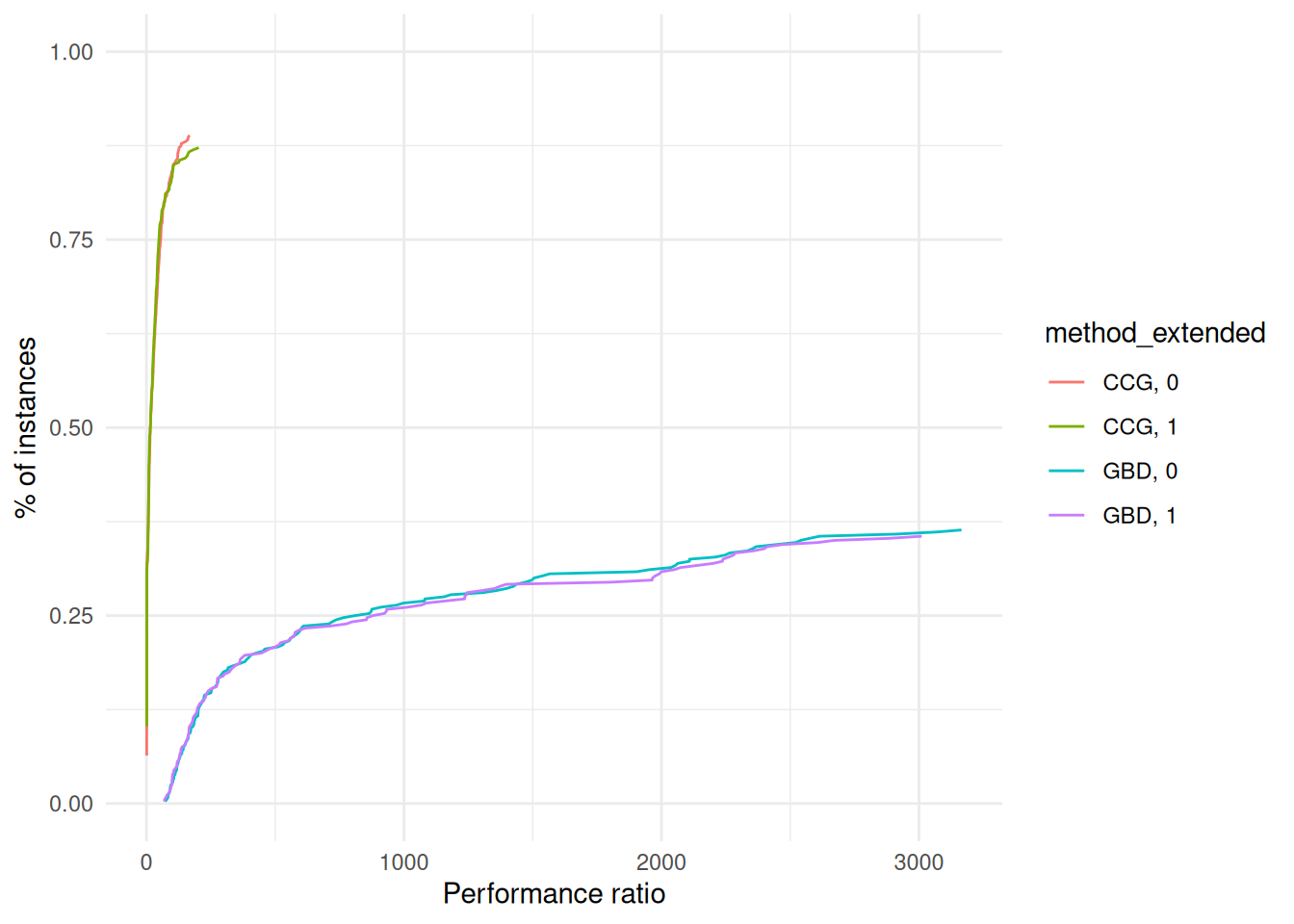
ggsave("performance_profile.pdf", plot = performance_profile)## Saving 7 x 5 in imageAs a complement, we also draw the ECDF.
ggplot(results, aes(x = total_time, color = method_extended)) +
stat_ecdf(geom = "step") +
xlab("Total Time") +
ylab("ECDF") +
labs(title = "ECDF of Total Time by Method") +
scale_x_continuous(breaks = seq(0, max(results$total_time), by = 500), labels = seq(0, max(results$total_time), by = 500)) +
theme_minimal()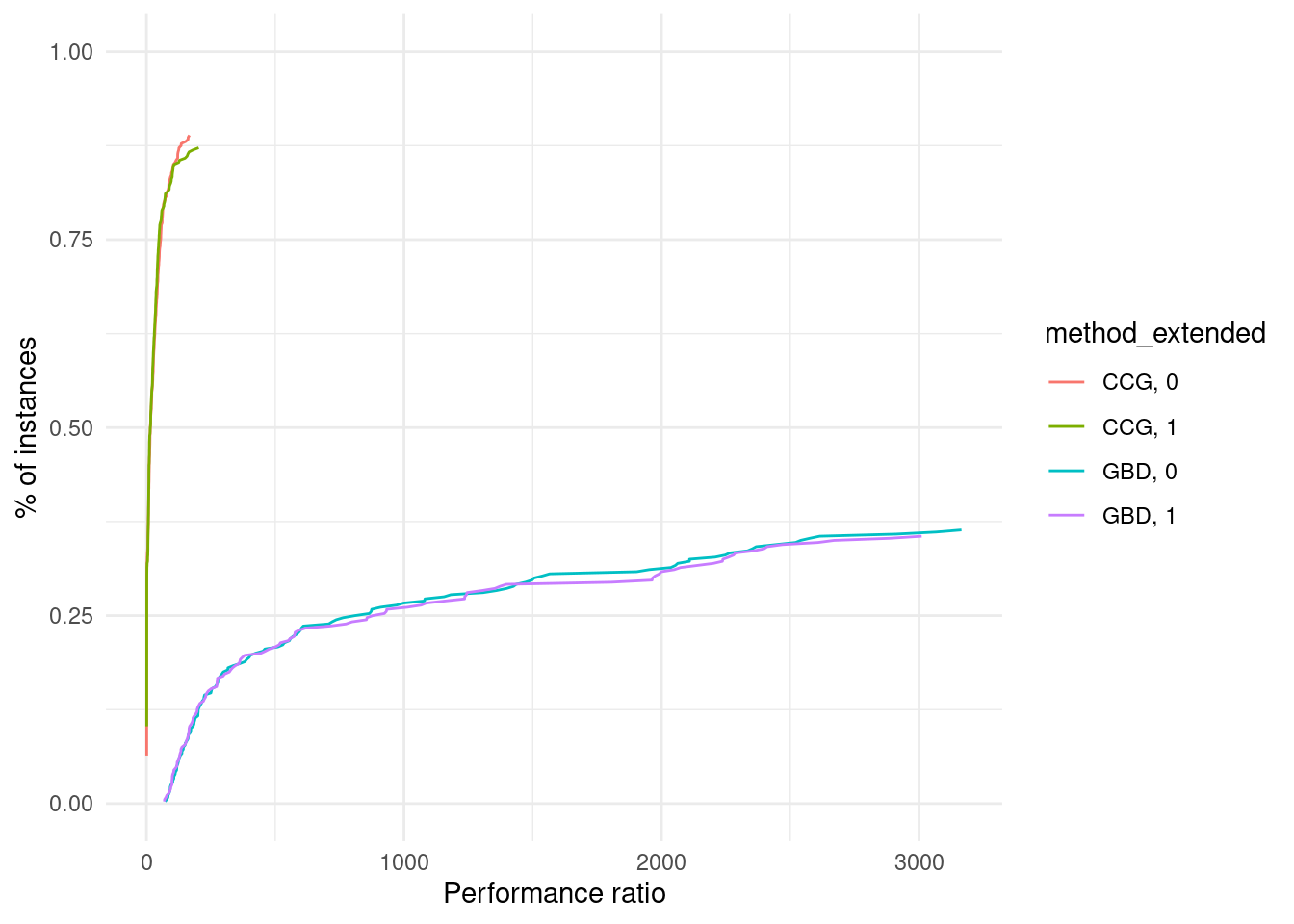
Summary table
In this section, we create a table summarizing the outcome of our experiments.
We start by computing average computation times over the solved instances.
results_solved = results %>%
filter(total_time < time_limit) %>%
group_by(method_extended, n_servers, n_clients, p, deviation) %>%
summarize(
solved = n(),
total_time = mean(total_time),
master_time = mean(master_time),
separation_time = mean(separation_time),
solved_iteration_count = mean(iteration_count),
.groups = "drop"
) %>%
ungroup()Then, we also consider unsolved instances: we count the number of such instances and compute the average iteration count.
results_unsolved <- results %>%
filter(total_time >= time_limit) %>%
group_by(method_extended, n_servers, n_clients, p, deviation) %>%
summarize(
unsolved = n(),
unsolved_iteration_count = mean(iteration_count),
.groups = "drop"
) %>%
ungroup()
results_fail <- results %>%
filter(fail == TRUE) %>%
group_by(method_extended, n_servers, n_clients, p, deviation) %>%
summarize(
fail = n(),
.groups = "drop"
) %>%
ungroup()We then merge the two tables.
final_result <- merge(results_solved, results_unsolved, by = c("method_extended", "n_servers", "n_clients", "p", "deviation"), all = TRUE)
final_result <- merge(final_result, results_fail, by = c("method_extended", "n_servers", "n_clients", "p", "deviation"), all = TRUE)Here is our table.
|
Solved instances
|
Unsolved instances
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | |V_1| | |V_2| | p | dev | Count | Total | Master | Sepatation | # Iter | Count | # Iter | # Errors |
| CCG, 0 | 10 | 20 | 0.1 | 0.25 | 10 | 2.76 | 0.07 | 2.64 | 2.80 | NA | NA | NA |
| CCG, 0 | 10 | 20 | 0.1 | 0.50 | 10 | 2.79 | 0.06 | 2.67 | 3.00 | NA | NA | NA |
| CCG, 0 | 10 | 20 | 0.2 | 0.25 | 10 | 20.97 | 0.06 | 20.85 | 3.00 | NA | NA | NA |
| CCG, 0 | 10 | 20 | 0.2 | 0.50 | 10 | 18.75 | 0.06 | 18.63 | 2.80 | NA | NA | NA |
| CCG, 0 | 10 | 20 | 0.3 | 0.25 | 10 | 64.48 | 0.08 | 64.32 | 3.00 | NA | NA | NA |
| CCG, 0 | 10 | 20 | 0.3 | 0.50 | 8 | 67.37 | 0.05 | 67.27 | 2.88 | 2 | 28.00 | 2 |
| CCG, 0 | 10 | 30 | 0.1 | 0.25 | 8 | 41.15 | 0.07 | 41.00 | 2.62 | 2 | 2.50 | 2 |
| CCG, 0 | 10 | 30 | 0.1 | 0.50 | 10 | 51.68 | 0.09 | 51.50 | 3.00 | NA | NA | NA |
| CCG, 0 | 10 | 30 | 0.2 | 0.25 | 9 | 1679.41 | 0.09 | 1679.23 | 3.11 | 1 | 1.00 | 1 |
| CCG, 0 | 10 | 30 | 0.2 | 0.50 | 9 | 1686.99 | 0.09 | 1686.81 | 3.11 | 1 | 2.00 | 1 |
| CCG, 0 | 10 | 30 | 0.3 | 0.25 | 4 | 3209.94 | 0.07 | 3209.79 | 2.50 | 6 | 1.83 | 2 |
| CCG, 0 | 10 | 30 | 0.3 | 0.50 | 4 | 3085.95 | 0.07 | 3085.81 | 2.50 | 6 | 1.67 | 1 |
| CCG, 0 | 15 | 20 | 0.1 | 0.25 | 10 | 3.60 | 0.08 | 3.44 | 2.80 | NA | NA | NA |
| CCG, 0 | 15 | 20 | 0.1 | 0.50 | 10 | 3.49 | 0.10 | 3.32 | 2.60 | NA | NA | NA |
| CCG, 0 | 15 | 20 | 0.2 | 0.25 | 10 | 30.09 | 0.09 | 29.91 | 2.80 | NA | NA | NA |
| CCG, 0 | 15 | 20 | 0.2 | 0.50 | 10 | 30.52 | 0.07 | 30.38 | 2.70 | NA | NA | NA |
| CCG, 0 | 15 | 20 | 0.3 | 0.25 | 10 | 138.92 | 0.11 | 138.73 | 3.00 | NA | NA | NA |
| CCG, 0 | 15 | 20 | 0.3 | 0.50 | 10 | 131.24 | 0.08 | 131.08 | 2.90 | NA | NA | NA |
| CCG, 0 | 15 | 30 | 0.1 | 0.25 | 10 | 50.36 | 0.13 | 50.11 | 3.00 | NA | NA | NA |
| CCG, 0 | 15 | 30 | 0.1 | 0.50 | 10 | 54.69 | 0.13 | 54.44 | 3.20 | NA | NA | NA |
| CCG, 0 | 15 | 30 | 0.2 | 0.25 | 10 | 1945.35 | 0.14 | 1945.08 | 3.50 | NA | NA | NA |
| CCG, 0 | 15 | 30 | 0.2 | 0.50 | 10 | 1916.98 | 0.13 | 1916.73 | 3.40 | NA | NA | NA |
| CCG, 0 | 15 | 30 | 0.3 | 0.25 | 6 | 4018.79 | 0.10 | 4018.58 | 3.17 | 4 | 1.50 | NA |
| CCG, 0 | 15 | 30 | 0.3 | 0.50 | 6 | 3568.58 | 0.11 | 3568.36 | 3.00 | 4 | 1.75 | NA |
| CCG, 0 | 20 | 20 | 0.1 | 0.25 | 10 | 4.34 | 0.11 | 4.14 | 2.80 | NA | NA | NA |
| CCG, 0 | 20 | 20 | 0.1 | 0.50 | 10 | 4.71 | 0.11 | 4.50 | 2.90 | NA | NA | NA |
| CCG, 0 | 20 | 20 | 0.2 | 0.25 | 10 | 37.34 | 0.10 | 37.14 | 2.90 | NA | NA | NA |
| CCG, 0 | 20 | 20 | 0.2 | 0.50 | 10 | 40.38 | 0.12 | 40.16 | 3.20 | NA | NA | NA |
| CCG, 0 | 20 | 20 | 0.3 | 0.25 | 10 | 120.73 | 0.10 | 120.53 | 3.00 | NA | NA | NA |
| CCG, 0 | 20 | 20 | 0.3 | 0.50 | 10 | 113.98 | 0.11 | 113.77 | 2.90 | NA | NA | NA |
| CCG, 0 | 20 | 30 | 0.1 | 0.25 | 10 | 65.89 | 0.11 | 65.65 | 2.70 | NA | NA | NA |
| CCG, 0 | 20 | 30 | 0.1 | 0.50 | 10 | 67.84 | 0.13 | 67.59 | 2.70 | NA | NA | NA |
| CCG, 0 | 20 | 30 | 0.2 | 0.25 | 10 | 2197.14 | 0.12 | 2196.89 | 2.80 | NA | NA | NA |
| CCG, 0 | 20 | 30 | 0.2 | 0.50 | 10 | 2179.94 | 0.13 | 2179.66 | 2.80 | NA | NA | NA |
| CCG, 0 | 20 | 30 | 0.3 | 0.25 | 3 | 4795.36 | 0.18 | 4795.05 | 2.67 | 7 | 1.86 | NA |
| CCG, 0 | 20 | 30 | 0.3 | 0.50 | 3 | 4519.99 | 0.13 | 4519.71 | 3.00 | 7 | 1.57 | NA |
| CCG, 1 | 10 | 20 | 0.1 | 0.25 | 10 | 2.64 | 0.13 | 2.42 | 4.50 | NA | NA | NA |
| CCG, 1 | 10 | 20 | 0.1 | 0.50 | 9 | 2.49 | 0.13 | 2.27 | 4.67 | 1 | 3.00 | 1 |
| CCG, 1 | 10 | 20 | 0.2 | 0.25 | 10 | 19.72 | 0.14 | 19.49 | 4.90 | NA | NA | NA |
| CCG, 1 | 10 | 20 | 0.2 | 0.50 | 10 | 17.30 | 0.12 | 17.11 | 4.70 | NA | NA | NA |
| CCG, 1 | 10 | 20 | 0.3 | 0.25 | 10 | 56.65 | 0.17 | 56.36 | 5.30 | NA | NA | NA |
| CCG, 1 | 10 | 20 | 0.3 | 0.50 | 9 | 56.27 | 0.13 | 56.06 | 4.89 | 1 | 7.00 | 1 |
| CCG, 1 | 10 | 30 | 0.1 | 0.25 | 8 | 38.82 | 0.19 | 38.51 | 5.12 | 2 | 1.00 | 2 |
| CCG, 1 | 10 | 30 | 0.1 | 0.50 | 8 | 44.02 | 0.18 | 43.72 | 5.00 | 2 | 7.00 | 2 |
| CCG, 1 | 10 | 30 | 0.2 | 0.25 | 8 | 1394.47 | 0.19 | 1394.15 | 4.88 | 2 | 2.50 | 2 |
| CCG, 1 | 10 | 30 | 0.2 | 0.50 | 8 | 1511.38 | 0.21 | 1511.04 | 5.12 | 2 | 2.00 | 2 |
| CCG, 1 | 10 | 30 | 0.3 | 0.25 | 3 | 3276.78 | 0.15 | 3276.52 | 4.33 | 7 | 3.29 | 2 |
| CCG, 1 | 10 | 30 | 0.3 | 0.50 | 2 | 3263.23 | 0.16 | 3262.95 | 4.50 | 8 | 3.62 | 5 |
| CCG, 1 | 15 | 20 | 0.1 | 0.25 | 10 | 3.34 | 0.18 | 3.04 | 4.70 | NA | NA | NA |
| CCG, 1 | 15 | 20 | 0.1 | 0.50 | 10 | 3.27 | 0.16 | 2.99 | 4.50 | NA | NA | NA |
| CCG, 1 | 15 | 20 | 0.2 | 0.25 | 10 | 26.58 | 0.18 | 26.28 | 4.70 | NA | NA | NA |
| CCG, 1 | 15 | 20 | 0.2 | 0.50 | 10 | 26.20 | 0.16 | 25.93 | 4.50 | NA | NA | NA |
| CCG, 1 | 15 | 20 | 0.3 | 0.25 | 10 | 116.75 | 0.22 | 116.40 | 5.20 | NA | NA | NA |
| CCG, 1 | 15 | 20 | 0.3 | 0.50 | 10 | 108.70 | 0.19 | 108.39 | 4.80 | NA | NA | NA |
| CCG, 1 | 15 | 30 | 0.1 | 0.25 | 10 | 44.96 | 0.28 | 44.51 | 5.00 | NA | NA | NA |
| CCG, 1 | 15 | 30 | 0.1 | 0.50 | 10 | 49.76 | 0.25 | 49.34 | 4.90 | NA | NA | NA |
| CCG, 1 | 15 | 30 | 0.2 | 0.25 | 9 | 1536.86 | 0.33 | 1536.34 | 5.67 | 1 | 6.00 | NA |
| CCG, 1 | 15 | 30 | 0.2 | 0.50 | 10 | 1677.56 | 0.33 | 1677.06 | 5.40 | NA | NA | NA |
| CCG, 1 | 15 | 30 | 0.3 | 0.25 | 7 | 3711.00 | 0.27 | 3710.57 | 4.71 | 3 | 3.67 | NA |
| CCG, 1 | 15 | 30 | 0.3 | 0.50 | 6 | 3476.93 | 0.25 | 3476.52 | 5.00 | 4 | 3.50 | NA |
| CCG, 1 | 20 | 20 | 0.1 | 0.25 | 10 | 4.11 | 0.22 | 3.75 | 4.70 | NA | NA | NA |
| CCG, 1 | 20 | 20 | 0.1 | 0.50 | 10 | 4.02 | 0.23 | 3.65 | 4.50 | NA | NA | NA |
| CCG, 1 | 20 | 20 | 0.2 | 0.25 | 10 | 29.47 | 0.24 | 29.08 | 4.80 | NA | NA | NA |
| CCG, 1 | 20 | 20 | 0.2 | 0.50 | 10 | 32.24 | 0.26 | 31.82 | 5.00 | NA | NA | NA |
| CCG, 1 | 20 | 20 | 0.3 | 0.25 | 10 | 107.48 | 0.26 | 107.06 | 5.10 | NA | NA | NA |
| CCG, 1 | 20 | 20 | 0.3 | 0.50 | 10 | 98.23 | 0.23 | 97.84 | 4.80 | NA | NA | NA |
| CCG, 1 | 20 | 30 | 0.1 | 0.25 | 10 | 53.45 | 0.27 | 52.99 | 4.50 | NA | NA | NA |
| CCG, 1 | 20 | 30 | 0.1 | 0.50 | 10 | 63.26 | 0.30 | 62.77 | 4.40 | NA | NA | NA |
| CCG, 1 | 20 | 30 | 0.2 | 0.25 | 10 | 1907.42 | 0.28 | 1906.94 | 4.70 | NA | NA | NA |
| CCG, 1 | 20 | 30 | 0.2 | 0.50 | 10 | 1937.70 | 0.31 | 1937.18 | 4.90 | NA | NA | NA |
| CCG, 1 | 20 | 30 | 0.3 | 0.25 | 3 | 3212.86 | 0.33 | 3212.34 | 4.33 | 7 | 4.00 | NA |
| CCG, 1 | 20 | 30 | 0.3 | 0.50 | 4 | 4247.61 | 0.33 | 4247.06 | 5.00 | 6 | 4.00 | NA |
| GBD, 0 | 10 | 20 | 0.1 | 0.25 | 10 | 260.65 | 0.51 | 259.88 | 269.40 | NA | NA | NA |
| GBD, 0 | 10 | 20 | 0.1 | 0.50 | 10 | 264.99 | 0.54 | 264.20 | 282.80 | NA | NA | NA |
| GBD, 0 | 10 | 20 | 0.2 | 0.25 | 10 | 2042.25 | 0.56 | 2041.38 | 283.40 | NA | NA | NA |
| GBD, 0 | 10 | 20 | 0.2 | 0.50 | 10 | 2013.00 | 0.53 | 2012.21 | 279.80 | NA | NA | NA |
| GBD, 0 | 10 | 20 | 0.3 | 0.25 | 5 | 3874.82 | 0.57 | 3873.97 | 275.20 | 5 | 227.00 | NA |
| GBD, 0 | 10 | 20 | 0.3 | 0.50 | 5 | 4102.41 | 0.57 | 4101.56 | 292.40 | 5 | 226.60 | NA |
| GBD, 0 | 10 | 30 | 0.1 | 0.25 | 10 | 5246.78 | 0.63 | 5245.81 | 289.10 | NA | NA | NA |
| GBD, 0 | 10 | 30 | 0.1 | 0.50 | 9 | 4958.93 | 0.60 | 4958.00 | 291.11 | 1 | 285.00 | NA |
| GBD, 0 | 10 | 30 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 24.20 | NA |
| GBD, 0 | 10 | 30 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 24.50 | NA |
| GBD, 0 | 10 | 30 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 10.20 | NA |
| GBD, 0 | 10 | 30 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 10.80 | NA |
| GBD, 0 | 15 | 20 | 0.1 | 0.25 | 10 | 671.14 | 1.71 | 668.87 | 517.80 | NA | NA | NA |
| GBD, 0 | 15 | 20 | 0.1 | 0.50 | 10 | 683.38 | 1.78 | 680.99 | 524.40 | NA | NA | NA |
| GBD, 0 | 15 | 20 | 0.2 | 0.25 | 8 | 5185.67 | 1.85 | 5183.23 | 547.75 | 2 | 369.50 | NA |
| GBD, 0 | 15 | 20 | 0.2 | 0.50 | 6 | 4432.99 | 1.70 | 4430.74 | 520.17 | 4 | 437.50 | NA |
| GBD, 0 | 15 | 20 | 0.3 | 0.25 | 1 | 4939.39 | 1.63 | 4937.23 | 552.00 | 9 | 229.89 | NA |
| GBD, 0 | 15 | 20 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 258.80 | NA |
| GBD, 0 | 15 | 30 | 0.1 | 0.25 | 1 | 4108.04 | 1.29 | 4106.23 | 473.00 | 9 | 400.11 | NA |
| GBD, 0 | 15 | 30 | 0.1 | 0.50 | 1 | 5399.63 | 1.73 | 5397.27 | 540.00 | 9 | 414.00 | NA |
| GBD, 0 | 15 | 30 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 24.10 | NA |
| GBD, 0 | 15 | 30 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 25.30 | NA |
| GBD, 0 | 15 | 30 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 12.20 | NA |
| GBD, 0 | 15 | 30 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 12.70 | 1 |
| GBD, 0 | 20 | 20 | 0.1 | 0.25 | 10 | 1311.56 | 5.42 | 1305.02 | 826.60 | NA | NA | NA |
| GBD, 0 | 20 | 20 | 0.1 | 0.50 | 10 | 1306.83 | 5.38 | 1300.35 | 818.80 | NA | NA | NA |
| GBD, 0 | 20 | 20 | 0.2 | 0.25 | 2 | 5201.39 | 4.32 | 5196.07 | 695.50 | 8 | 557.75 | NA |
| GBD, 0 | 20 | 20 | 0.2 | 0.50 | 3 | 5371.14 | 4.16 | 5366.07 | 745.67 | 7 | 524.43 | NA |
| GBD, 0 | 20 | 20 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 253.70 | NA |
| GBD, 0 | 20 | 20 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 243.20 | NA |
| GBD, 0 | 20 | 30 | 0.1 | 0.25 | NA | NA | NA | NA | NA | 10 | 314.80 | NA |
| GBD, 0 | 20 | 30 | 0.1 | 0.50 | NA | NA | NA | NA | NA | 10 | 308.60 | NA |
| GBD, 0 | 20 | 30 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 13.20 | NA |
| GBD, 0 | 20 | 30 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 12.00 | NA |
| GBD, 0 | 20 | 30 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 4.90 | NA |
| GBD, 0 | 20 | 30 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 5.20 | NA |
| GBD, 1 | 10 | 20 | 0.1 | 0.25 | 9 | 231.08 | 0.63 | 230.15 | 314.78 | 1 | 87904.00 | 1 |
| GBD, 1 | 10 | 20 | 0.1 | 0.50 | 9 | 251.93 | 0.69 | 250.93 | 339.67 | 1 | 87085.00 | 1 |
| GBD, 1 | 10 | 20 | 0.2 | 0.25 | 8 | 1743.32 | 1.00 | 1741.92 | 419.38 | 2 | 85708.00 | 2 |
| GBD, 1 | 10 | 20 | 0.2 | 0.50 | 10 | 1905.25 | 1.10 | 1903.73 | 445.00 | NA | NA | NA |
| GBD, 1 | 10 | 20 | 0.3 | 0.25 | 5 | 3906.72 | 1.04 | 3905.27 | 414.40 | 5 | 35926.40 | 2 |
| GBD, 1 | 10 | 20 | 0.3 | 0.50 | 7 | 4637.00 | 1.39 | 4635.16 | 511.57 | 3 | 517.33 | NA |
| GBD, 1 | 10 | 30 | 0.1 | 0.25 | 9 | 4630.08 | 0.79 | 4628.89 | 344.78 | 1 | 86737.00 | 1 |
| GBD, 1 | 10 | 30 | 0.1 | 0.50 | 10 | 5020.28 | 0.63 | 5019.31 | 298.00 | NA | NA | NA |
| GBD, 1 | 10 | 30 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 26556.90 | 3 |
| GBD, 1 | 10 | 30 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 17632.10 | 2 |
| GBD, 1 | 10 | 30 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 147.80 | NA |
| GBD, 1 | 10 | 30 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 27812.10 | 3 |
| GBD, 1 | 15 | 20 | 0.1 | 0.25 | 10 | 628.54 | 3.71 | 623.97 | 738.30 | NA | NA | NA |
| GBD, 1 | 15 | 20 | 0.1 | 0.50 | 10 | 632.16 | 3.08 | 628.31 | 731.10 | NA | NA | NA |
| GBD, 1 | 15 | 20 | 0.2 | 0.25 | 7 | 4364.68 | 3.24 | 4360.68 | 778.43 | 3 | 598.00 | NA |
| GBD, 1 | 15 | 20 | 0.2 | 0.50 | 5 | 4689.21 | 4.62 | 4683.69 | 882.80 | 5 | 16987.20 | 1 |
| GBD, 1 | 15 | 20 | 0.3 | 0.25 | 1 | 4839.30 | 3.55 | 4834.61 | 861.00 | 9 | 557.78 | NA |
| GBD, 1 | 15 | 20 | 0.3 | 0.50 | 1 | 5655.09 | 4.31 | 5649.87 | 940.00 | 9 | 577.33 | NA |
| GBD, 1 | 15 | 30 | 0.1 | 0.25 | 2 | 5309.26 | 2.59 | 5305.88 | 701.50 | 8 | 28807.88 | 3 |
| GBD, 1 | 15 | 30 | 0.1 | 0.50 | 1 | 4855.06 | 1.80 | 4852.61 | 592.00 | 9 | 525.56 | NA |
| GBD, 1 | 15 | 30 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 7530.90 | 1 |
| GBD, 1 | 15 | 30 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 23269.30 | 3 |
| GBD, 1 | 15 | 30 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 7415.90 | 1 |
| GBD, 1 | 15 | 30 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 15543.40 | 2 |
| GBD, 1 | 20 | 20 | 0.1 | 0.25 | 10 | 1216.71 | 9.87 | 1205.41 | 1102.10 | NA | NA | NA |
| GBD, 1 | 20 | 20 | 0.1 | 0.50 | 9 | 1272.45 | 10.21 | 1260.65 | 1159.11 | 1 | 68967.00 | 1 |
| GBD, 1 | 20 | 20 | 0.2 | 0.25 | 2 | 4697.51 | 5.30 | 4691.15 | 819.00 | 8 | 1044.50 | NA |
| GBD, 1 | 20 | 20 | 0.2 | 0.50 | 3 | 5650.41 | 17.13 | 5631.38 | 1336.67 | 7 | 9322.57 | 1 |
| GBD, 1 | 20 | 20 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 793.60 | NA |
| GBD, 1 | 20 | 20 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 692.10 | NA |
| GBD, 1 | 20 | 30 | 0.1 | 0.25 | NA | NA | NA | NA | NA | 10 | 5863.50 | 1 |
| GBD, 1 | 20 | 30 | 0.1 | 0.50 | NA | NA | NA | NA | NA | 10 | 474.70 | NA |
| GBD, 1 | 20 | 30 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 392.50 | NA |
| GBD, 1 | 20 | 30 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 13984.70 | 2 |
| GBD, 1 | 20 | 30 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 13890.40 | 2 |
| GBD, 1 | 20 | 30 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 13481.50 | 2 |
Revision
Reading Additional Results for the Revision
We start by reading the file and by removing the “tag” column.
results_rev = read.table("./results-rev.csv", header = FALSE, sep = ',')
colnames(results_rev) <- c("tag", "instance", "n_servers", "n_clients", "Gamma", "deviation", "time_limit", "method", "use_heuristic", "total_time", "master_time", "separation_time", "best_bound", "iteration_count", "fail", "use_bilevel_separation", "use_budgeted_unc")results = read.table("./new-results.csv", header = FALSE, sep = ',')
colnames(results) <- c("tag", "instance", "n_servers", "n_clients", "Gamma", "deviation", "time_limit", "method", "use_heuristic", "total_time", "master_time", "separation_time", "best_bound", "iteration_count", "fail")
results$use_bilevel_separation = 0
results$use_budgeted_unc = 1all_results = rbind(results, results_rev)
all_results = all_results %>% filter(method == "CCG")Then, we compute the percentage which \(\Gamma\) represents for the total number of clients.
all_results$p = all_results$Gamma / all_results$n_clients
all_results$p = ceiling(all_results$p / .05) * .05all_results$size = paste0("(", all_results$n_servers, ",", all_results$n_clients, "), ", all_results$p)all_results$method_extended = paste0(all_results$method, ", ", ifelse(all_results$use_heuristic, "heuristic, ", ""), ifelse(all_results$use_bilevel_separation, "KKT", "0-1 mapping"))
paged_table(results_rev %>% filter(fail == 1 & total_time < time_limit))paged_table(results_rev %>% filter(fail == 0 & deviation == .25))if (sum(all_results$fail) > 0) {
all_results[all_results$fail == TRUE,]$total_time = time_limit
}
all_results$unsolved = all_results$total_time >= time_limit | all_results$failbudgeted_unc_results = all_results %>% filter(use_budgeted_unc == 1 & method == "CCG")
ggplot(budgeted_unc_results, aes(x = total_time, color = method_extended)) +
stat_ecdf(geom = "step", linewidth = 1) +
labs(title = "ECDF Plot", x = "Values", y = "ECDF") +
theme_minimal()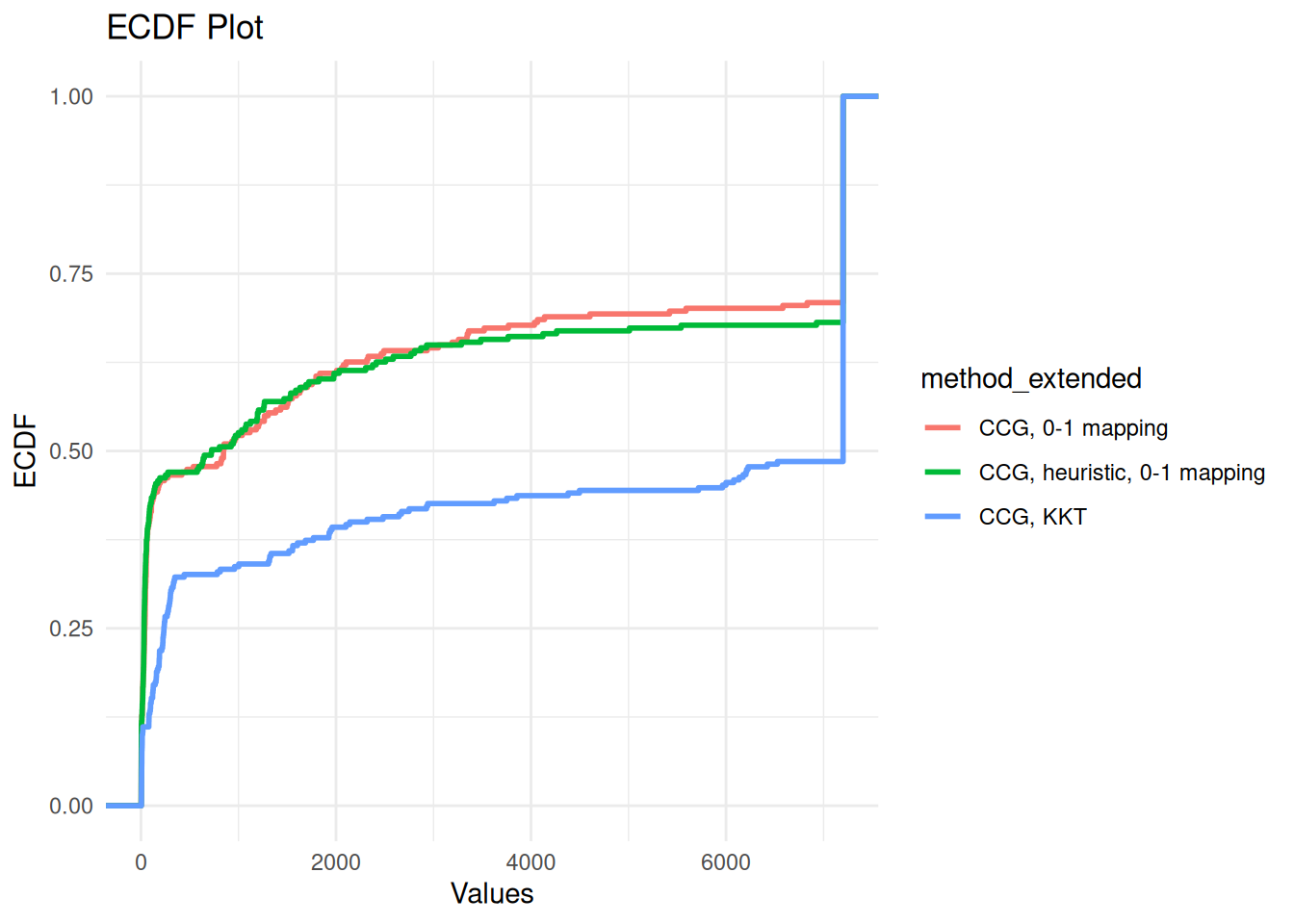
performance_profile = plot_performance_profile(budgeted_unc_results, criterion_column = "total_time", solver_column = "method_extended") +
labs(x = "Performance ratio", y = "% of instances") +
scale_y_continuous(limits = c(0, 1)) +
theme_minimal()
print(performance_profile)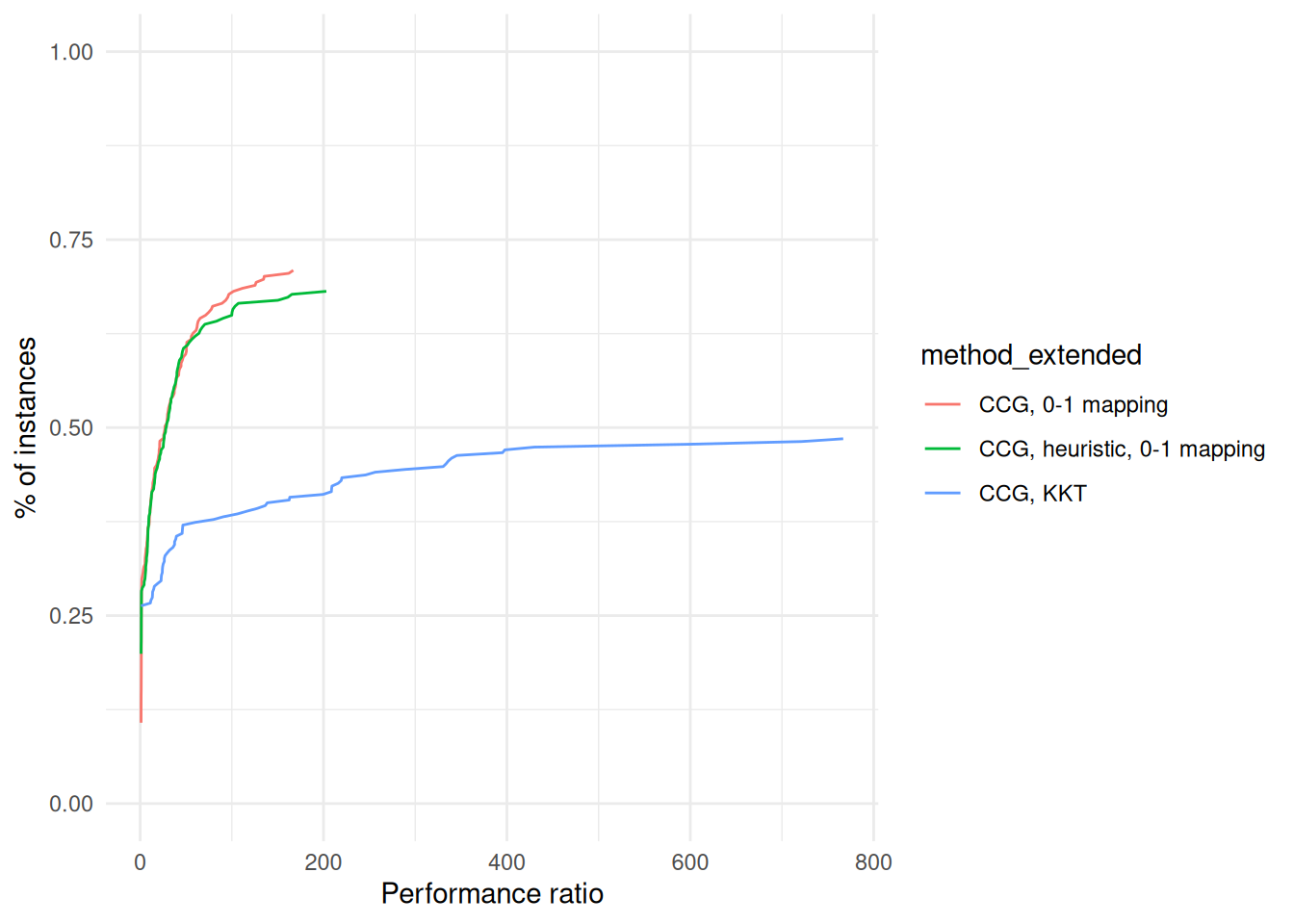
knapsack_unc_results = all_results %>% filter(method == "CCG" & use_budgeted_unc == 0)
ggplot(knapsack_unc_results, aes(x = total_time, color = method_extended)) +
stat_ecdf(geom = "step", size = 1) +
labs(title = "ECDF Plot", x = "Values", y = "ECDF") +
facet_wrap(~p) +
theme_minimal()## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
## ℹ Please use `linewidth` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.
Summary table
In this section, we create a table summarizing the outcome of our experiments.
We start by computing average computation times over the solved instances.
results_solved = knapsack_unc_results %>%
filter(total_time < time_limit) %>%
group_by(method_extended, n_servers, n_clients, p, deviation) %>%
summarize(
solved = n(),
total_time = mean(total_time),
master_time = mean(master_time),
separation_time = mean(separation_time),
solved_iteration_count = mean(iteration_count),
.groups = "drop"
) %>%
ungroup()Then, we also consider unsolved instances: we count the number of such instances and compute the average iteration count.
results_unsolved <- knapsack_unc_results %>%
filter(total_time >= time_limit) %>%
group_by(method_extended, n_servers, n_clients, p, deviation) %>%
summarize(
unsolved = n(),
unsolved_iteration_count = mean(iteration_count),
.groups = "drop"
) %>%
ungroup()
results_fail <- knapsack_unc_results %>%
filter(fail == TRUE) %>%
group_by(method_extended, n_servers, n_clients, p, deviation) %>%
summarize(
fail = n(),
.groups = "drop"
) %>%
ungroup()We then merge the two tables.
final_result <- merge(results_solved, results_unsolved, by = c("method_extended", "n_servers", "n_clients", "p", "deviation"), all = TRUE)
final_result <- merge(final_result, results_fail, by = c("method_extended", "n_servers", "n_clients", "p", "deviation"), all = TRUE)Here is our table.
|
Solved instances
|
Unsolved instances
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | |V_1| | |V_2| | p | dev | Count | Total | Master | Sepatation | # Iter | Count | # Iter | # Errors |
| CCG, KKT | 10 | 20 | 0.1 | 0.25 | 10 | 1238.42 | 0.10 | 1238.27 | 2.50 | NA | NA | NA |
| CCG, KKT | 10 | 20 | 0.1 | 0.50 | 10 | 2316.82 | 0.13 | 2316.64 | 2.80 | NA | NA | NA |
| CCG, KKT | 10 | 20 | 0.2 | 0.25 | 3 | 4382.48 | 0.21 | 4382.19 | 4.00 | 7 | 1.71 | NA |
| CCG, KKT | 10 | 20 | 0.2 | 0.50 | 1 | 4680.02 | 0.17 | 4679.75 | 5.00 | 9 | 1.33 | NA |
| CCG, KKT | 10 | 20 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 1.50 | NA |
| CCG, KKT | 10 | 20 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 1.00 | NA |
| CCG, KKT | 10 | 30 | 0.1 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.10 | NA |
| CCG, KKT | 10 | 30 | 0.1 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.10 | NA |
| CCG, KKT | 10 | 30 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 10 | 30 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 10 | 30 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 10 | 30 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 10 | 40 | 0.1 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 10 | 40 | 0.1 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 10 | 40 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 10 | 40 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 10 | 40 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 10 | 40 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 15 | 20 | 0.1 | 0.25 | 8 | 2776.59 | 0.07 | 2776.46 | 3.00 | 2 | 1.50 | NA |
| CCG, KKT | 15 | 20 | 0.1 | 0.50 | 8 | 2524.01 | 0.09 | 2523.86 | 3.12 | 2 | 1.50 | NA |
| CCG, KKT | 15 | 20 | 0.2 | 0.25 | 2 | 4688.58 | 0.07 | 4688.45 | 3.00 | 8 | 1.75 | NA |
| CCG, KKT | 15 | 20 | 0.2 | 0.50 | 2 | 6671.90 | 0.07 | 6671.78 | 3.00 | 8 | 0.88 | NA |
| CCG, KKT | 15 | 20 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 1.10 | NA |
| CCG, KKT | 15 | 20 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 1.20 | NA |
| CCG, KKT | 15 | 30 | 0.1 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.30 | NA |
| CCG, KKT | 15 | 30 | 0.1 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.20 | NA |
| CCG, KKT | 15 | 30 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.10 | NA |
| CCG, KKT | 15 | 30 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 15 | 30 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 15 | 30 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 15 | 40 | 0.1 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 15 | 40 | 0.1 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 15 | 40 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 15 | 40 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 15 | 40 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 15 | 40 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 20 | 20 | 0.1 | 0.25 | 6 | 3058.98 | 0.11 | 3058.79 | 3.33 | 4 | 2.50 | NA |
| CCG, KKT | 20 | 20 | 0.1 | 0.50 | 7 | 4030.51 | 0.11 | 4030.33 | 3.29 | 3 | 2.00 | NA |
| CCG, KKT | 20 | 20 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 1.10 | NA |
| CCG, KKT | 20 | 20 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 1.30 | NA |
| CCG, KKT | 20 | 20 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 1.10 | NA |
| CCG, KKT | 20 | 20 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 1.00 | NA |
| CCG, KKT | 20 | 30 | 0.1 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.20 | NA |
| CCG, KKT | 20 | 30 | 0.1 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.10 | NA |
| CCG, KKT | 20 | 30 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 20 | 30 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 20 | 30 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 20 | 30 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 20 | 40 | 0.1 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 20 | 40 | 0.1 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 20 | 40 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 20 | 40 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 20 | 40 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
| CCG, KKT | 20 | 40 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 0.00 | NA |
Comparison with Static Robust Problem
static = read.table("./static-rap-results.csv", header = FALSE, sep = ',')
colnames(static) <- c("tag", "instance", "n_servers", "n_clients", "Gamma", "deviation", "time_limit", "method", "use_heuristic", "total_time", "master_time", "separation_time", "best_bound", "iteration_count", "fail", "use_bilevel_separation", "use_budgeted_unc")
static$tag = NULL columns_to_keep = c("instance", "Gamma", "deviation", "n_servers", "n_clients", "method", "total_time", "best_bound", "iteration_count")
ccg = all_results %>% filter(method == "CCG", use_heuristic == 0, use_bilevel_separation == 0, use_budgeted_unc == 1) %>% select(all_of(columns_to_keep), p)
static = static %>% select(columns_to_keep)## Warning: Using an external vector in selections was deprecated in tidyselect 1.1.0.
## ℹ Please use `all_of()` or `any_of()` instead.
## # Was:
## data %>% select(columns_to_keep)
##
## # Now:
## data %>% select(all_of(columns_to_keep))
##
## See <https://tidyselect.r-lib.org/reference/faq-external-vector.html>.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.ccg = ccg %>% mutate(instance = basename(instance))
static = static %>% mutate(instance = basename(instance))
merged_df = merge(ccg, static,
by = c("instance", "Gamma", "deviation"),
all = TRUE,
suffixes = c(".ccg", ".static"))
merged_df %>% filter(best_bound.ccg > best_bound.static)## instance Gamma deviation n_servers.ccg n_clients.ccg
## 1 instance_S10_C20__2.txt 6 0.50 10 20
## 2 instance_S10_C20__4.txt 6 0.50 10 20
## 3 instance_S10_C30__0.txt 3 0.25 10 30
## 4 instance_S10_C30__0.txt 6 0.25 10 30
## 5 instance_S10_C30__0.txt 6 0.50 10 30
## 6 instance_S10_C30__0.txt 9 0.25 10 30
## 7 instance_S10_C30__0.txt 9 0.50 10 30
## 8 instance_S10_C30__7.txt 3 0.25 10 30
## 9 instance_S10_C30__9.txt 9 0.25 10 30
## 10 instance_S10_C40__0.txt 4 0.50 10 40
## 11 instance_S10_C40__1.txt 4 0.25 10 40
## 12 instance_S10_C40__1.txt 4 0.50 10 40
## 13 instance_S10_C40__2.txt 4 0.25 10 40
## 14 instance_S10_C40__2.txt 4 0.50 10 40
## 15 instance_S10_C40__2.txt 8 0.50 10 40
## 16 instance_S10_C40__3.txt 4 0.50 10 40
## 17 instance_S10_C40__4.txt 4 0.25 10 40
## 18 instance_S10_C40__4.txt 4 0.50 10 40
## 19 instance_S10_C40__6.txt 4 0.25 10 40
## 20 instance_S10_C40__6.txt 4 0.50 10 40
## 21 instance_S10_C40__7.txt 4 0.25 10 40
## 22 instance_S10_C40__7.txt 4 0.50 10 40
## 23 instance_S10_C40__8.txt 4 0.50 10 40
## 24 instance_S10_C40__9.txt 4 0.25 10 40
## 25 instance_S15_C40__8.txt 4 0.50 15 40
## method.ccg total_time.ccg best_bound.ccg iteration_count.ccg p
## 1 CCG 7200 1e+20 2 0.3
## 2 CCG 7200 1e+20 54 0.3
## 3 CCG 7200 1e+20 2 0.1
## 4 CCG 7200 1e+20 1 0.2
## 5 CCG 7200 1e+20 2 0.2
## 6 CCG 7200 1e+20 3 0.3
## 7 CCG 7200 1e+20 2 0.3
## 8 CCG 7200 1e+20 3 0.1
## 9 CCG 7200 1e+20 2 0.3
## 10 CCG 7200 1e+20 1 0.1
## 11 CCG 7200 1e+20 1 0.1
## 12 CCG 7200 1e+20 1 0.1
## 13 CCG 7200 1e+20 1 0.1
## 14 CCG 7200 1e+20 2 0.1
## 15 CCG 7200 1e+20 1 0.2
## 16 CCG 7200 1e+20 2 0.1
## 17 CCG 7200 1e+20 1 0.1
## 18 CCG 7200 1e+20 1 0.1
## 19 CCG 7200 1e+20 2 0.1
## 20 CCG 7200 1e+20 2 0.1
## 21 CCG 7200 1e+20 3 0.1
## 22 CCG 7200 1e+20 2 0.1
## 23 CCG 7200 1e+20 3 0.1
## 24 CCG 7200 1e+20 2 0.1
## 25 CCG 7200 1e+20 1 0.1
## n_servers.static n_clients.static method.static total_time.static
## 1 10 20 Static 0.073676
## 2 10 20 Static 0.017307
## 3 10 30 Static 0.017379
## 4 10 30 Static 0.015327
## 5 10 30 Static 0.012198
## 6 10 30 Static 0.017425
## 7 10 30 Static 0.013167
## 8 10 30 Static 0.078152
## 9 10 30 Static 0.075932
## 10 10 40 Static 0.074957
## 11 10 40 Static 0.016686
## 12 10 40 Static 0.017202
## 13 10 40 Static 0.080372
## 14 10 40 Static 0.081235
## 15 10 40 Static 0.081203
## 16 10 40 Static 0.078905
## 17 10 40 Static 0.084900
## 18 10 40 Static 0.086383
## 19 10 40 Static 0.336255
## 20 10 40 Static 0.239991
## 21 10 40 Static 0.068940
## 22 10 40 Static 0.068841
## 23 10 40 Static 0.076108
## 24 10 40 Static 0.063384
## 25 15 40 Static 0.081368
## best_bound.static iteration_count.static
## 1 1129810 0
## 2 1000650 0
## 3 2154810 0
## 4 2154810 0
## 5 3088660 0
## 6 2154810 0
## 7 3088660 0
## 8 1273510 0
## 9 1974460 0
## 10 3634090 0
## 11 6488470 0
## 12 9326280 0
## 13 3835130 0
## 14 5507930 0
## 15 5507930 0
## 16 4386170 0
## 17 5312360 0
## 18 7632320 0
## 19 3464220 0
## 20 4973340 0
## 21 2407820 0
## 22 3450730 0
## 23 3408460 0
## 24 2441350 0
## 25 3531820 0merged_df %>% filter(best_bound.static < 1e-4)## [1] instance Gamma deviation
## [4] n_servers.ccg n_clients.ccg method.ccg
## [7] total_time.ccg best_bound.ccg iteration_count.ccg
## [10] p n_servers.static n_clients.static
## [13] method.static total_time.static best_bound.static
## [16] iteration_count.static
## <0 rows> (or 0-length row.names)library(tidyr)
# 1. Compute summary
summary_table <- merged_df %>%
mutate(
gap = (best_bound.static - best_bound.ccg) / (1e-10 + abs(best_bound.ccg)) * 100,
time_status = ifelse(total_time.ccg < time_limit, "within_time", "timed_out")
) %>%
group_by(n_servers.ccg, n_clients.ccg, p, deviation, time_status) %>%
summarise(
n_instances = length(best_bound.static),
n_missing_static = sum(is.na(best_bound.static)),
avg_ccg_time = mean(total_time.ccg),
avg_static_time = mean(total_time.static),
avg_gap = ifelse(sum(!is.na(best_bound.static) & best_bound.ccg < 1e20) > 0,
mean(gap[!is.na(best_bound.static) & best_bound.ccg < 1e20]),
NA),
.groups = "drop"
)
# 2. Pivot wider
summary_wide <- summary_table %>%
pivot_wider(
names_from = time_status,
values_from = c(n_instances, n_missing_static, avg_ccg_time, avg_static_time, avg_gap),
names_glue = "{time_status}_{.value}"
) %>%
arrange(n_servers.ccg, n_clients.ccg, p, deviation)
summary_wide = summary_wide %>% select(n_servers.ccg, n_clients.ccg, p, deviation, within_time_n_instances, within_time_n_missing_static, within_time_avg_ccg_time, within_time_avg_static_time, within_time_avg_gap, timed_out_n_instances, timed_out_n_missing_static, timed_out_avg_gap)
# 3. Pretty table with exact column names
kable(summary_wide, "html", digits = 4, escape = FALSE, col.names = c(
"|V_1|", "|V_2|", "p", "dev",
"n_instances", "n_static_is_infeasible", "avg_ccg_time", "avg_static_time", "avg_gap (%)",
"n_instances", "n_static_is_infeasible", "avg_gap (%)"
)) %>%
kable_styling(full_width = FALSE, position = "center") %>%
add_header_above(c(" " = 4, "Solved by CCG" = 5, "Time out" = 3))| |V_1| | |V_2| | p | dev | n_instances | n_static_is_infeasible | avg_ccg_time | avg_static_time | avg_gap (%) | n_instances | n_static_is_infeasible | avg_gap (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 20 | 0.1 | 0.25 | 10 | 0 | 2.7630 | 0.0516 | 39.7193 | NA | NA | NA |
| 10 | 20 | 0.1 | 0.50 | 10 | 0 | 2.7936 | 0.0746 | 81.0962 | NA | NA | NA |
| 10 | 20 | 0.2 | 0.25 | 10 | 0 | 20.9713 | 0.0484 | 29.3203 | NA | NA | NA |
| 10 | 20 | 0.2 | 0.50 | 10 | 0 | 18.7494 | 0.0708 | 56.6216 | NA | NA | NA |
| 10 | 20 | 0.3 | 0.25 | 10 | 0 | 64.4844 | 0.0361 | 21.2249 | NA | NA | NA |
| 10 | 20 | 0.3 | 0.50 | 8 | 0 | 67.3693 | 0.0685 | 39.8887 | 2 | 0 | NA |
| 10 | 30 | 0.1 | 0.25 | 8 | 0 | 41.1517 | 0.1045 | 40.2544 | 2 | 0 | NA |
| 10 | 30 | 0.1 | 0.50 | 10 | 0 | 51.6795 | 0.1006 | 82.1599 | NA | NA | NA |
| 10 | 30 | 0.2 | 0.25 | 9 | 0 | 1679.4066 | 0.1005 | 29.8757 | 1 | 0 | NA |
| 10 | 30 | 0.2 | 0.50 | 9 | 0 | 1686.9894 | 0.1014 | 57.9487 | 1 | 0 | NA |
| 10 | 30 | 0.3 | 0.25 | 4 | 0 | 3209.9375 | 0.1323 | 20.8472 | 6 | 0 | 2.382870e+01 |
| 10 | 30 | 0.3 | 0.50 | 4 | 0 | 3085.9475 | 0.1352 | 38.0109 | 6 | 0 | 4.422800e+01 |
| 10 | 40 | 0.1 | 0.25 | 4 | 0 | 851.0570 | 0.1021 | 40.4610 | 6 | 0 | NA |
| 10 | 40 | 0.1 | 0.50 | 2 | 0 | 1423.9850 | 0.0737 | 85.7246 | 8 | 0 | NA |
| 10 | 40 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 0 | 2.690682e+18 |
| 10 | 40 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 0 | 4.290233e+18 |
| 10 | 40 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 0 | 3.420617e+18 |
| 10 | 40 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 0 | 4.909337e+18 |
| 15 | 20 | 0.1 | 0.25 | 10 | 0 | 3.5984 | 0.0368 | 39.1436 | NA | NA | NA |
| 15 | 20 | 0.1 | 0.50 | 10 | 0 | 3.4870 | 0.0495 | 80.0390 | NA | NA | NA |
| 15 | 20 | 0.2 | 0.25 | 10 | 0 | 30.0851 | 0.0247 | 29.1977 | NA | NA | NA |
| 15 | 20 | 0.2 | 0.50 | 10 | 0 | 30.5192 | 0.0312 | 56.5144 | NA | NA | NA |
| 15 | 20 | 0.3 | 0.25 | 10 | 0 | 138.9164 | 0.0569 | 21.5904 | NA | NA | NA |
| 15 | 20 | 0.3 | 0.50 | 10 | 0 | 131.2409 | 0.0571 | 40.1713 | NA | NA | NA |
| 15 | 30 | 0.1 | 0.25 | 10 | 0 | 50.3552 | 0.0631 | 39.0970 | NA | NA | NA |
| 15 | 30 | 0.1 | 0.50 | 10 | 0 | 54.6920 | 0.0629 | 79.7976 | NA | NA | NA |
| 15 | 30 | 0.2 | 0.25 | 10 | 0 | 1945.3498 | 0.0559 | 28.7398 | NA | NA | NA |
| 15 | 30 | 0.2 | 0.50 | 10 | 0 | 1916.9835 | 0.0564 | 55.5301 | NA | NA | NA |
| 15 | 30 | 0.3 | 0.25 | 6 | 0 | 4018.7925 | 0.0634 | 19.8371 | 4 | 0 | 2.279190e+01 |
| 15 | 30 | 0.3 | 0.50 | 6 | 0 | 3568.5817 | 0.0432 | 36.7971 | 4 | 0 | 4.250240e+01 |
| 15 | 40 | 0.1 | 0.25 | 10 | 0 | 1519.0398 | 0.0919 | 38.8737 | NA | NA | NA |
| 15 | 40 | 0.1 | 0.50 | 9 | 0 | 1264.2533 | 0.0911 | 79.0396 | 1 | 0 | NA |
| 15 | 40 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 10 | 0 | 1.903920e+18 |
| 15 | 40 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 10 | 0 | 2.718815e+18 |
| 15 | 40 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 10 | 0 | 2.079132e+18 |
| 15 | 40 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 10 | 0 | 2.968791e+18 |
| 20 | 20 | 0.1 | 0.25 | 10 | 0 | 4.3418 | 0.0734 | 36.6646 | NA | NA | NA |
| 20 | 20 | 0.1 | 0.50 | 10 | 0 | 4.7058 | 0.0766 | 74.6413 | NA | NA | NA |
| 20 | 20 | 0.2 | 0.25 | 10 | 0 | 37.3417 | 0.0990 | 27.1353 | NA | NA | NA |
| 20 | 20 | 0.2 | 0.50 | 10 | 0 | 40.3835 | 0.0740 | 52.5028 | NA | NA | NA |
| 20 | 20 | 0.3 | 0.25 | 10 | 0 | 120.7265 | 0.0752 | 19.7950 | NA | NA | NA |
| 20 | 20 | 0.3 | 0.50 | 10 | 0 | 113.9756 | 0.0958 | 36.7518 | NA | NA | NA |
| 20 | 30 | 0.1 | 0.25 | 10 | 0 | 65.8871 | 0.0928 | 38.3800 | NA | NA | NA |
| 20 | 30 | 0.1 | 0.50 | 10 | 0 | 67.8437 | 0.0827 | 78.2346 | NA | NA | NA |
| 20 | 30 | 0.2 | 0.25 | 10 | 0 | 2197.1450 | 0.0863 | 28.1594 | NA | NA | NA |
| 20 | 30 | 0.2 | 0.50 | 10 | 0 | 2179.9431 | 0.0687 | 54.3321 | NA | NA | NA |
| 20 | 30 | 0.3 | 0.25 | 3 | 0 | 4795.3567 | 0.0743 | 19.1452 | 7 | 0 | 2.131300e+01 |
| 20 | 30 | 0.3 | 0.50 | 3 | 0 | 4519.9900 | 0.0738 | 34.3281 | 7 | 0 | 4.020530e+01 |
| 20 | 40 | 0.1 | 0.25 | 4 | 0 | 1294.6280 | 0.0821 | 39.9275 | NA | NA | NA |
| 20 | 40 | 0.1 | 0.50 | 4 | 0 | 1570.1735 | 0.0822 | 81.7340 | NA | NA | NA |
| 20 | 40 | 0.2 | 0.25 | NA | NA | NA | NA | NA | 4 | 0 | 2.216242e+18 |
| 20 | 40 | 0.2 | 0.50 | NA | NA | NA | NA | NA | 4 | 0 | 3.167708e+18 |
| 20 | 40 | 0.3 | 0.25 | NA | NA | NA | NA | NA | 3 | 0 | 2.954990e+18 |
| 20 | 40 | 0.3 | 0.50 | NA | NA | NA | NA | NA | 3 | 0 | 4.223610e+18 |
| NA | NA | NA | 0.25 | NA | NA | NA | NA | NA | NA | NA | NA |
| NA | NA | NA | 0.50 | NA | NA | NA | NA | NA | NA | NA | NA |
Larger instances
larger = read.table("./larger-results.csv", header = FALSE, sep = ',')
colnames(larger) <- c("tag", "instance", "n_servers", "n_clients", "Gamma", "deviation", "time_limit", "method", "use_heuristic", "total_time", "master_time", "separation_time", "best_bound", "iteration_count", "fail", "use_bilevel_separation", "use_budgeted_unc")
larger$tag = NULL
larger$p = larger$Gamma / larger$n_clients
larger$p = ceiling(larger$p / .05) * .05
ccg = larger %>% filter(method == "CCG", use_heuristic == 1)
static = larger %>% filter(method == "Static")
static$p = NULL
merged_df = merge(ccg, static,
by = c("instance", "Gamma", "deviation"),
all = TRUE,
suffixes = c(".ccg", ".static"))
merged_df %>% filter(best_bound.ccg > best_bound.static)## [1] instance Gamma
## [3] deviation n_servers.ccg
## [5] n_clients.ccg time_limit.ccg
## [7] method.ccg use_heuristic.ccg
## [9] total_time.ccg master_time.ccg
## [11] separation_time.ccg best_bound.ccg
## [13] iteration_count.ccg fail.ccg
## [15] use_bilevel_separation.ccg use_budgeted_unc.ccg
## [17] p n_servers.static
## [19] n_clients.static time_limit.static
## [21] method.static use_heuristic.static
## [23] total_time.static master_time.static
## [25] separation_time.static best_bound.static
## [27] iteration_count.static fail.static
## [29] use_bilevel_separation.static use_budgeted_unc.static
## <0 rows> (or 0-length row.names)merged_df %>% filter(best_bound.static < 1e-4)## [1] instance Gamma
## [3] deviation n_servers.ccg
## [5] n_clients.ccg time_limit.ccg
## [7] method.ccg use_heuristic.ccg
## [9] total_time.ccg master_time.ccg
## [11] separation_time.ccg best_bound.ccg
## [13] iteration_count.ccg fail.ccg
## [15] use_bilevel_separation.ccg use_budgeted_unc.ccg
## [17] p n_servers.static
## [19] n_clients.static time_limit.static
## [21] method.static use_heuristic.static
## [23] total_time.static master_time.static
## [25] separation_time.static best_bound.static
## [27] iteration_count.static fail.static
## [29] use_bilevel_separation.static use_budgeted_unc.static
## <0 rows> (or 0-length row.names)# 1. Compute summary
summary_table <- merged_df %>%
mutate(
gap = (best_bound.static - best_bound.ccg) / (1e-10 + abs(best_bound.ccg)) * 100,
time_status = "timed_out",#ifelse(total_time.ccg < time_limit, "within_time", "timed_out")
) %>%
group_by(n_servers.ccg, n_clients.ccg, p, deviation, time_status) %>%
summarise(
n_instances = length(best_bound.static),
n_missing_static = sum(is.na(best_bound.static)),
avg_ccg_time = mean(total_time.ccg),
avg_static_time = mean(total_time.static),
avg_gap = ifelse(sum(!is.na(best_bound.static) & best_bound.ccg < 1e20) > 0,
mean(gap[!is.na(best_bound.static) & best_bound.ccg < 1e20]),
NA),
.groups = "drop"
)
# 2. Pivot wider
summary_wide <- summary_table %>%
pivot_wider(
names_from = time_status,
values_from = c(n_instances, n_missing_static, avg_ccg_time, avg_static_time, avg_gap),
names_glue = "{time_status}_{.value}"
) %>%
arrange(n_servers.ccg, n_clients.ccg, p, deviation)
summary_wide = summary_wide %>% select(n_servers.ccg, n_clients.ccg, p, deviation, timed_out_n_instances, timed_out_n_missing_static, timed_out_avg_gap)
# 3. Pretty table with exact column names
kable(summary_wide, "html", digits = 4, escape = FALSE, col.names = c(
"|V_1|", "|V_2|", "p", "dev",
"n_instances", "n_static_is_infeasible", "avg_gap (%)"
)) %>%
kable_styling(full_width = FALSE, position = "center") %>%
add_header_above(c(" " = 4, "Time out" = 3))| |V_1| | |V_2| | p | dev | n_instances | n_static_is_infeasible | avg_gap (%) |
|---|---|---|---|---|---|---|
| 25 | 30 | 0.1 | 0.25 | 10 | 0 | 39.1086 |
| 25 | 30 | 0.1 | 0.50 | 10 | 0 | 80.1877 |
| 25 | 30 | 0.2 | 0.25 | 10 | 0 | 29.0000 |
| 25 | 30 | 0.2 | 0.50 | 10 | 0 | 56.2415 |
| 25 | 30 | 0.3 | 0.25 | 10 | 0 | 21.1857 |
| 25 | 30 | 0.3 | 0.50 | 10 | 0 | 39.3478 |
| 25 | 40 | 0.1 | 0.25 | 10 | 0 | 38.8186 |
| 25 | 40 | 0.1 | 0.50 | 10 | 0 | 79.3348 |
| 25 | 40 | 0.2 | 0.25 | 10 | 0 | 29.6624 |
| 25 | 40 | 0.2 | 0.50 | 10 | 0 | 56.6800 |
| 25 | 40 | 0.3 | 0.25 | 10 | 0 | 21.8574 |
| 25 | 40 | 0.3 | 0.50 | 10 | 0 | 40.0643 |
| 30 | 30 | 0.1 | 0.25 | 10 | 0 | 36.5976 |
| 30 | 30 | 0.1 | 0.50 | 10 | 0 | 74.6052 |
| 30 | 30 | 0.2 | 0.25 | 10 | 0 | 27.4052 |
| 30 | 30 | 0.2 | 0.50 | 10 | 0 | 53.2447 |
| 30 | 30 | 0.3 | 0.25 | 10 | 0 | 20.7143 |
| 30 | 30 | 0.3 | 0.50 | 10 | 0 | 38.0615 |
| 30 | 40 | 0.1 | 0.25 | 10 | 0 | 38.3789 |
| 30 | 40 | 0.1 | 0.50 | 10 | 0 | 78.5781 |
| 30 | 40 | 0.2 | 0.25 | 10 | 0 | 29.0527 |
| 30 | 40 | 0.2 | 0.50 | 10 | 0 | 56.4982 |
| 30 | 40 | 0.3 | 0.25 | 10 | 0 | 21.7515 |
| 30 | 40 | 0.3 | 0.50 | 10 | 0 | 40.4267 |
git push action on the public repository
hlefebvr/hlefebvr.github.io using rmarkdown and Github
Actions. This ensures the reproducibility of our data manipulation. The
last compilation was performed on the 20/01/26 18:19:57.
 Henri Lefebvre
Henri Lefebvre