Table of Contents
PADM for Mxied-integer Bilevel Optimization
export_data_from_ggplot_ecdf = function(plot, xmax = NULL) {
plot_data = ggplot_build(plot)$data[[1]]
plot_data = plot_data %>% filter(is.finite(x))
if (is.null(xmax)) {
xmax = max(plot_data$x, na.rm = TRUE)
}
ecdf_wide = plot_data %>%
select(x, y, group) %>%
pivot_wider(names_from = group, values_from = y) %>%
arrange(x) %>%
slice(1:n()-1)
final_row = ecdf_wide %>%
select(-x) %>%
summarise(across(everything(), ~ max(.x, na.rm = TRUE))) %>%
mutate(x = xmax) %>%
select(x, everything())
ecdf_wide = bind_rows(ecdf_wide, final_row)
return(ecdf_wide)
}
export_data_from_ggplot_ecdf_csv = function(plot, filename) {
result = export_data_from_ggplot_ecdf(plot)
write.csv(result, file = filename, na = "")
}Loading BOBILib
bobilib_columns = c("instance", "class", "type", "n_upper_vars", "n_upper_integer_vars", "n_upper_binary_vars", "n_lower_vars", "n_lower_integer_vars", "n_lower_binary_vars", "n_linking_vars", "n_linking_integer_vars", "n_linking_binary_vars", "n_linking_continuous_vars", "n_upper_ctrs", "n_lower_ctrs", "n_coupling_ctrs", "best_known_obj", "bobilib_status", "difficulty")
bobilib = read.csv("bobilib.csv", header = FALSE, col.names = bobilib_columns)
bobilib = bobilib %>% mutate(n_vars = as.numeric(n_upper_vars + n_lower_vars),
n_ctrs = as.numeric(n_upper_ctrs + n_lower_ctrs))
paged_table(bobilib)Loading Results
# Function to read results obtained on the HPC
get_results_from_hpc = function(method) {
result = read.csv(paste0("results/results-", method, ".csv"), header = FALSE, sep = ",")
if (method == "PADM") {
colnames(result) = c("tag", "instance", "method", "update_rule", "factor", "initial_penalty_parameter", "time", "status", "reason", "best_obj", "outer_loops", "inner_loops", "lambda", "pi_lb", "pi_ub")
result[result$status != "Feasible",]$time = 3600
result = result %>% mutate(method = paste0(method, "-", initial_penalty_parameter))
} else if (method == "PADM-interdiction") {
colnames(result) = c("tag", "instance", "method", "update_rule", "factor", "initial_penalty_parameter", "time", "status", "reason", "best_obj", "outer_loops", "inner_loops")
result[result$status != "Feasible",]$time = 3600
result = result %>% mutate(method = paste0(method, "-", initial_penalty_parameter))
} else {
colnames(result) = c("tag", "instance", "method", "time", "status", "reason", "best_bound", "best_obj", "lambda", "pi_lb", "pi_ub")
}
result = result %>% mutate(instance = basename(instance), instance = gsub("\\.aux$", "", instance))
result = result %>% mutate(time = ifelse(time > 3600, 3600, time))
return(result)
}
# Function to read results obtained by Johannes for BOBILib
get_results_from_boblib = function(method) {
result = read.csv(paste0("results-bobilib/", method, ".csv"), header = FALSE, sep = ",")
colnames(result) = c("instance", "method", "time", "best_obj")
result = result %>% mutate(time = ifelse(time > 3600, 3600, time))
return(result)
}
# Reading all results
columns = c("instance", "method", "time", "best_obj")
mibs = get_results_from_boblib("MibS") %>% select(all_of(columns))
filemosi = get_results_from_boblib("filemosi") %>% select(all_of(columns))
milp = get_results_from_hpc("MILP") %>% select(all_of(columns))
milp_first = get_results_from_hpc("MILP-first") %>% select(all_of(columns))
#nlp = get_results_from_hpc("NLP") %>% select(all_of(columns))
padm = get_results_from_hpc("PADM") %>% select(all_of(columns))
padm_interdiction = get_results_from_hpc("PADM-interdiction") %>% select(all_of(columns))
# Selecting only those results fulfilling our assumptions
instances = unique(unlist(lapply(list(milp,padm), function(df) df$instance)))
add_missing_results = function(t_data) {
result = t_data
missing_instances = setdiff(instances, t_data$instance)
if (length(missing_instances) > 0) {
method = unique(t_data$method)
missing_results = data.frame(
instance = missing_instances,
method = method,
time = 3600,
best_obj = NA
)
result = rbind(result, missing_results)
}
result = result %>% filter(instance %in% instances)
return(result)
}
mibs = add_missing_results(mibs)
filemosi = add_missing_results(filemosi)
milp = add_missing_results(milp)
#nlp = add_missing_results(nlp)
padm = add_missing_results(padm)
milp_first = add_missing_results(milp_first)
# Aggregating all results
all_results = rbind(
mibs, milp, milp_first, padm, padm_interdiction
)
all_results = all_results %>%
left_join(bobilib %>% select(instance, bobilib_status, best_known_obj, class, type, n_vars, n_ctrs), by = "instance") %>%
mutate(
best_known_obj = na_if(best_known_obj, "None"),
best_obj = na_if(best_obj, "None")
) %>%
mutate(
best_known_obj = as.numeric(best_known_obj),
best_obj = as.numeric(best_obj)
)
paged_table(all_results)all_results %>%
distinct(instance, .keep_all = TRUE) %>%
count(bobilib_status)## bobilib_status n
## 1 infeasible 29
## 2 open 285
## 3 open with feasible point 1132
## 4 optimal 770all_results %>%
distinct(instance, .keep_all = TRUE) %>%
count(class, type)## class type n
## 1 general-bilevel mixed-integer 166
## 2 general-bilevel pure-integer 92
## 3 interdiction assignment 24
## 4 interdiction clique 219
## 5 interdiction generalized 90
## 6 interdiction knapsack 599
## 7 interdiction multidimensional-knapsack 954
## 8 interdiction network 72PADM
Computational Times
# Step 0: Compute size per unique instance and assign quantile bins as factors
instance_sizes <- all_results %>%
distinct(instance, n_vars, n_ctrs) %>%
mutate(
size = n_vars + n_ctrs,
size_group = ntile(size, 5),
size_group = factor(size_group) # convert to factor here
)
# Step 1: Create readable interval labels for each bin, keeping size_group as factor
group_labels <- instance_sizes %>%
group_by(size_group) %>%
summarise(
min_size = min(size),
max_size = max(size),
.groups = "drop"
) %>%
mutate(
label = paste0(min_size, "--", max_size),
size_group = factor(size_group) # factor again for safety
)
# Step 2: Join size and group info into main data (filter methods first)
all_results_with_size <- all_results %>%
filter(method %in% c("MibS", "MILP", "MILP-first", "PADM-1000")) %>%
left_join(instance_sizes %>% select(instance, size_group, size), by = "instance") %>%
left_join(group_labels, by = "size_group") %>%
mutate(size_group_label = factor(label, levels = group_labels$label)) # preserve order
# Step 3: Count total instances per group (unique problems)
total_counts <- all_results_with_size %>%
distinct(instance, size_group_label) %>%
count(size_group_label, name = "count") %>%
mutate(type = "Total")
# Step 4: Count solved instances per method
solved_counts <- all_results_with_size %>%
filter(time < 3600) %>%
group_by(size_group_label, method) %>%
summarise(count = n(), .groups = "drop") %>%
rename(type = method)
# Step 4.1: Fill in missing combinations (show 0 count bars)
all_methods <- unique(all_results_with_size$method)
all_groups <- unique(all_results_with_size$size_group_label)
solved_counts <- solved_counts %>%
complete(size_group_label = all_groups, type = all_methods, fill = list(count = 0))
# Step 5: Combine and plot
plot_data <- bind_rows(total_counts, solved_counts)
ggplot(plot_data, aes(x = size_group_label, y = count, fill = type)) +
geom_bar(stat = "identity", position = "dodge") +
labs(
title = "Instance and Solved Counts by Size Interval",
x = "Size Interval",
y = "Count",
fill = "Type"
) +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1))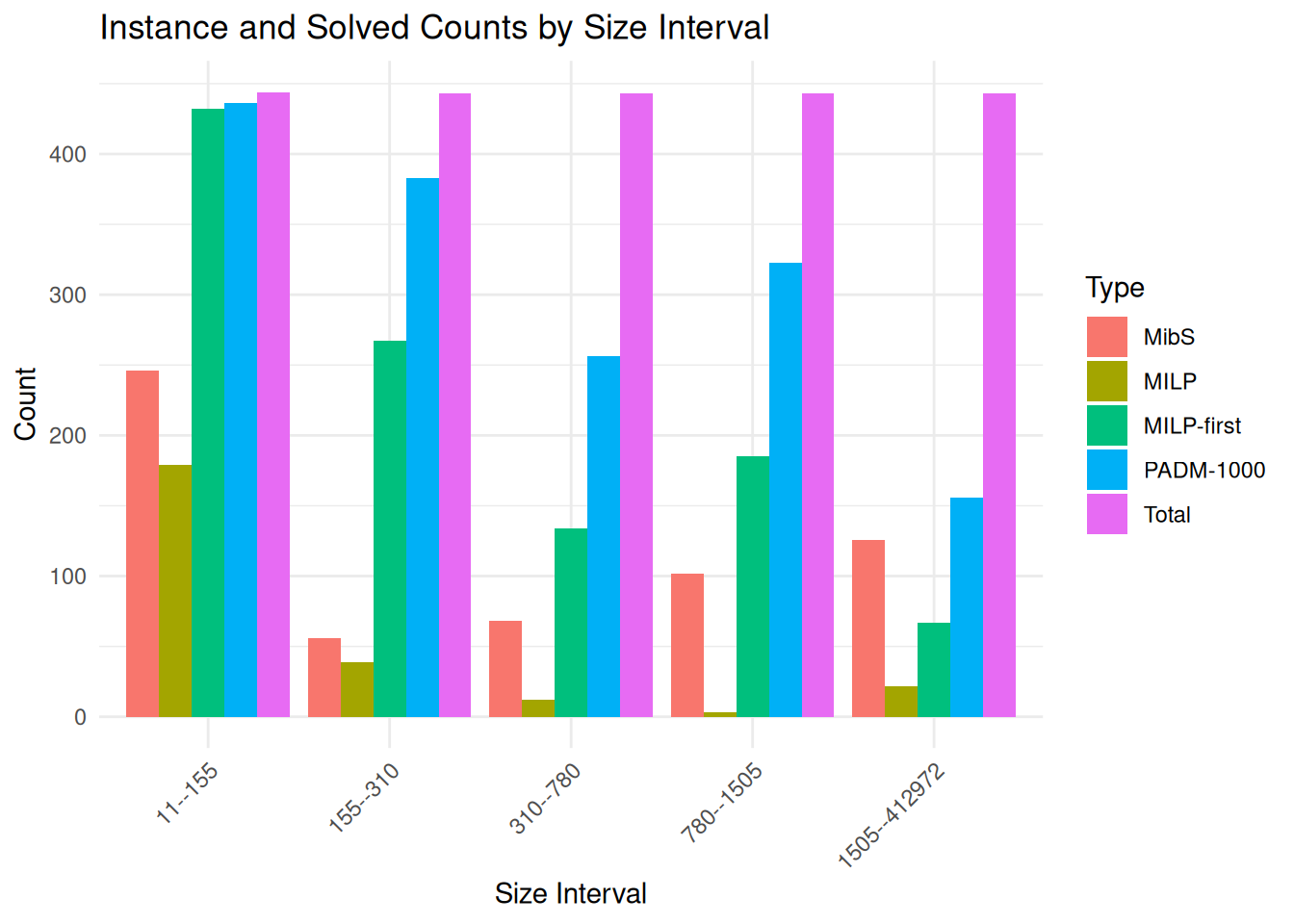
wide_data = plot_data %>%
pivot_wider(
names_from = type,
values_from = count,
values_fill = 0 # Fill missing combinations with 0
)
#write.csv(wide_data, "barplot_solved.csv", row.names = FALSE, quote = FALSE)plot_ecdf_of_time = function(data, log = FALSE) {
p = ggplot(data, aes(x = time, color = method)) +
stat_ecdf(geom = "step", linewidth = 1) +
labs(
title = "ECDF of Time",
x = "Time (s)",
y = "Instances (%)"
) +
theme_minimal() +
scale_color_brewer(palette = "Set1")
if (log) {
p = p + scale_x_log10()
}
return(p)
}
p = plot_ecdf_of_time(all_results %>% filter(!startsWith(method, "PADM-interdiction")))
p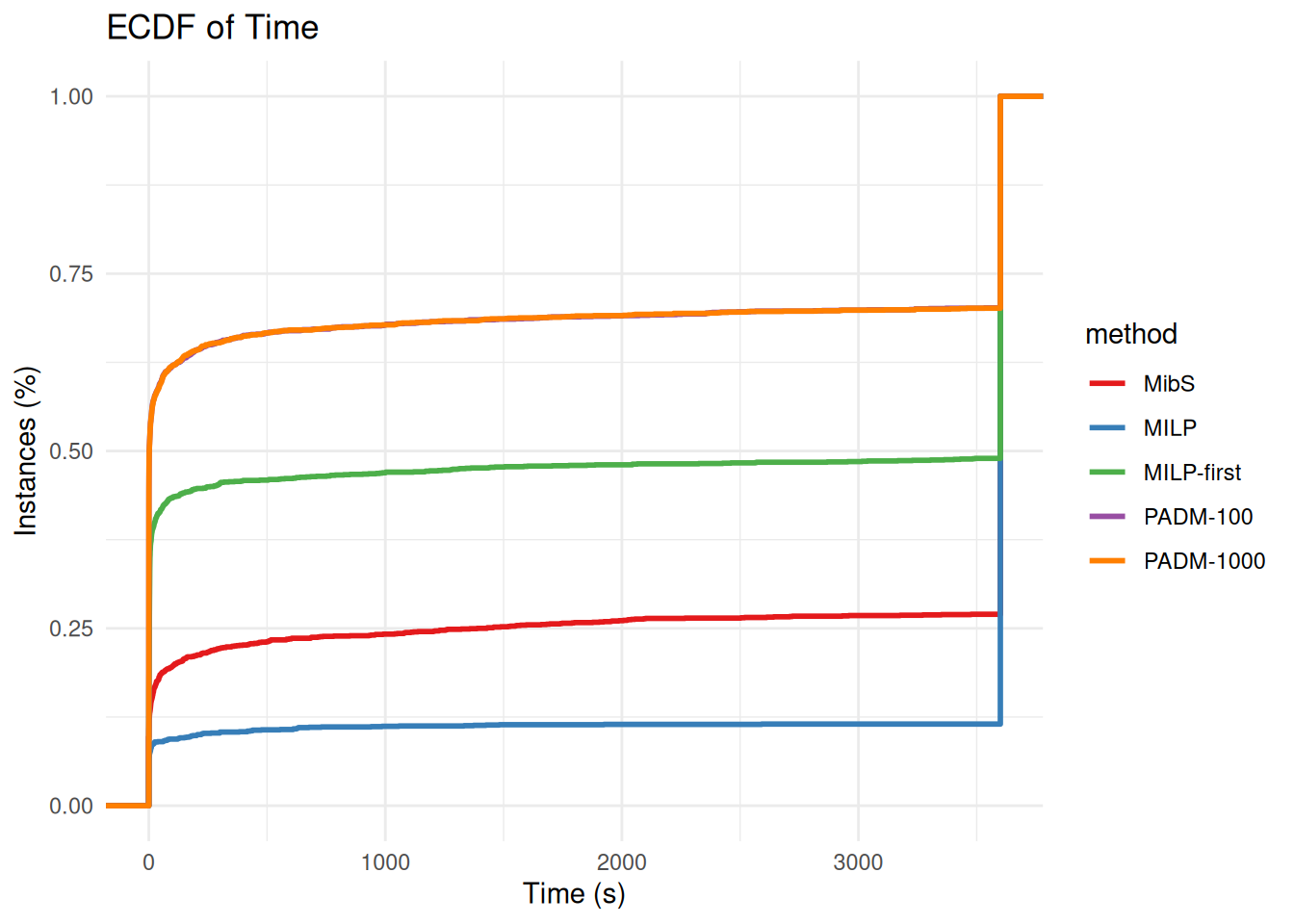
export_data_from_ggplot_ecdf_csv(p, "ecdf_time_all.csv")padm_raw = get_results_from_hpc("PADM") %>% filter(method == "PADM-100")
ggplot(padm_raw, aes(x = lambda, color = method)) +
stat_ecdf(geom = "step", linewidth = 1) +
scale_x_log10() +
labs(
title = "ECDF of Lambda bound",
x = "Value",
y = "Instances (%)"
) +
theme_minimal() +
scale_color_brewer(palette = "Set1") 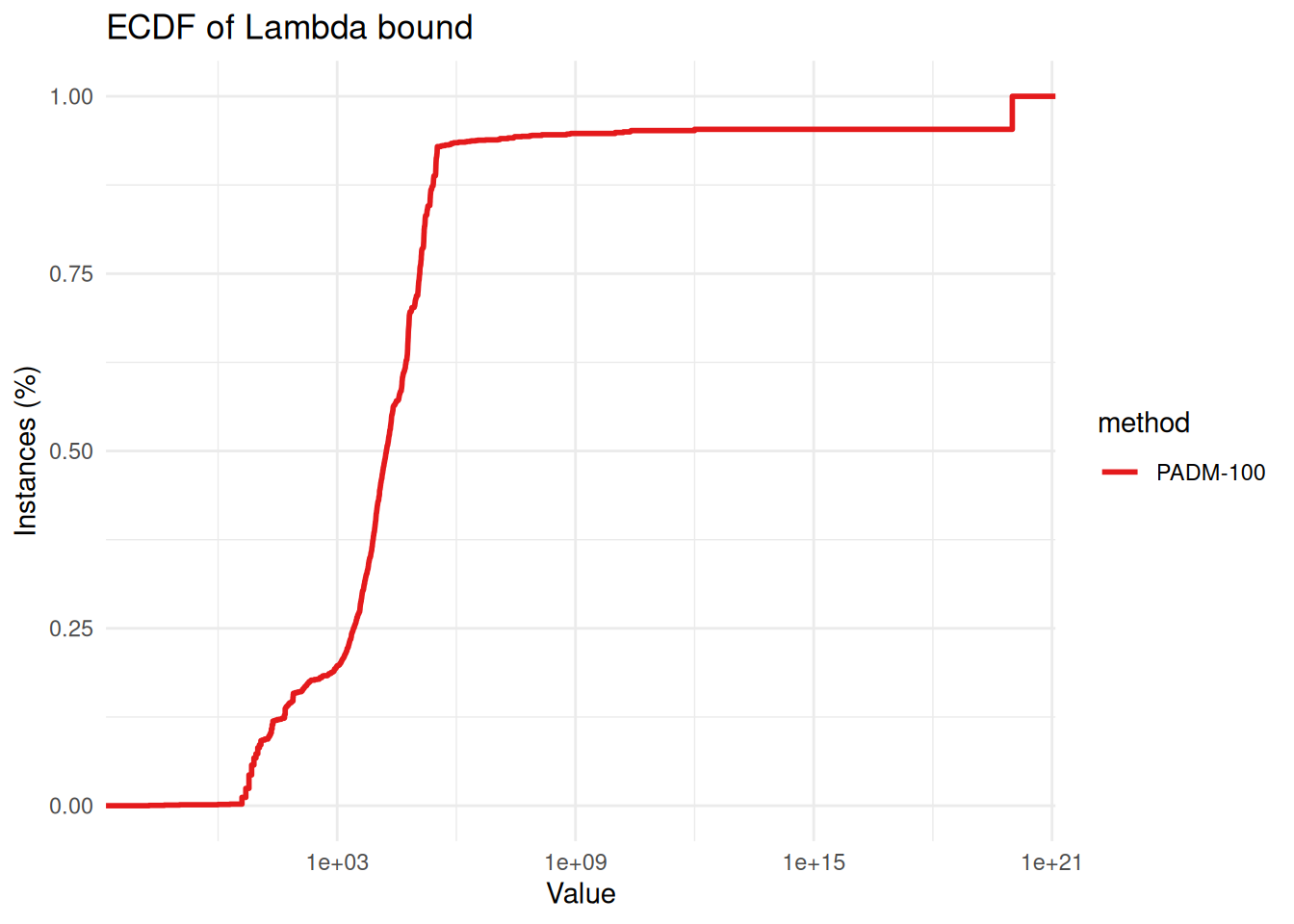
summary(padm_raw$lambda)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.000e+00 2.582e+03 1.727e+04 4.650e+18 1.196e+05 1.000e+20# padm_raw %>% filter(lambda > 1e10)Optimality Gap
plot_ecdf_of_gap_wrt_best_known = function(data) {
results_with_gaps = data %>%
filter(!is.na(best_known_obj)) %>%
filter(bobilib_status == "optimal") %>%
filter(time < 3600) %>%
mutate(gap = abs(best_obj - best_known_obj) / (1e-10 + abs(best_known_obj))) %>%
mutate(gap = ifelse(gap < 1e-4, 1e-4, gap))
#print(results_with_gaps %>% filter(method == "MILP-first" & gap > .8 & gap < 1.2 ))
p = ggplot(results_with_gaps, aes(x = gap, color = method)) +
stat_ecdf(geom = "step", linewidth = 1) +
#scale_x_log10() +
coord_cartesian(xlim = c(1e-4, 2e1)) +
labs(
title = "ECDF of Gap",
x = "Gap (%)",
y = "Instances (%)"
) +
theme_minimal() +
scale_color_brewer(palette = "Set1")
return(p)
}
plot_ecdf_of_gap_wrt_best_known(all_results %>% filter(method == "PADM-100" | method == "PADM-1000"))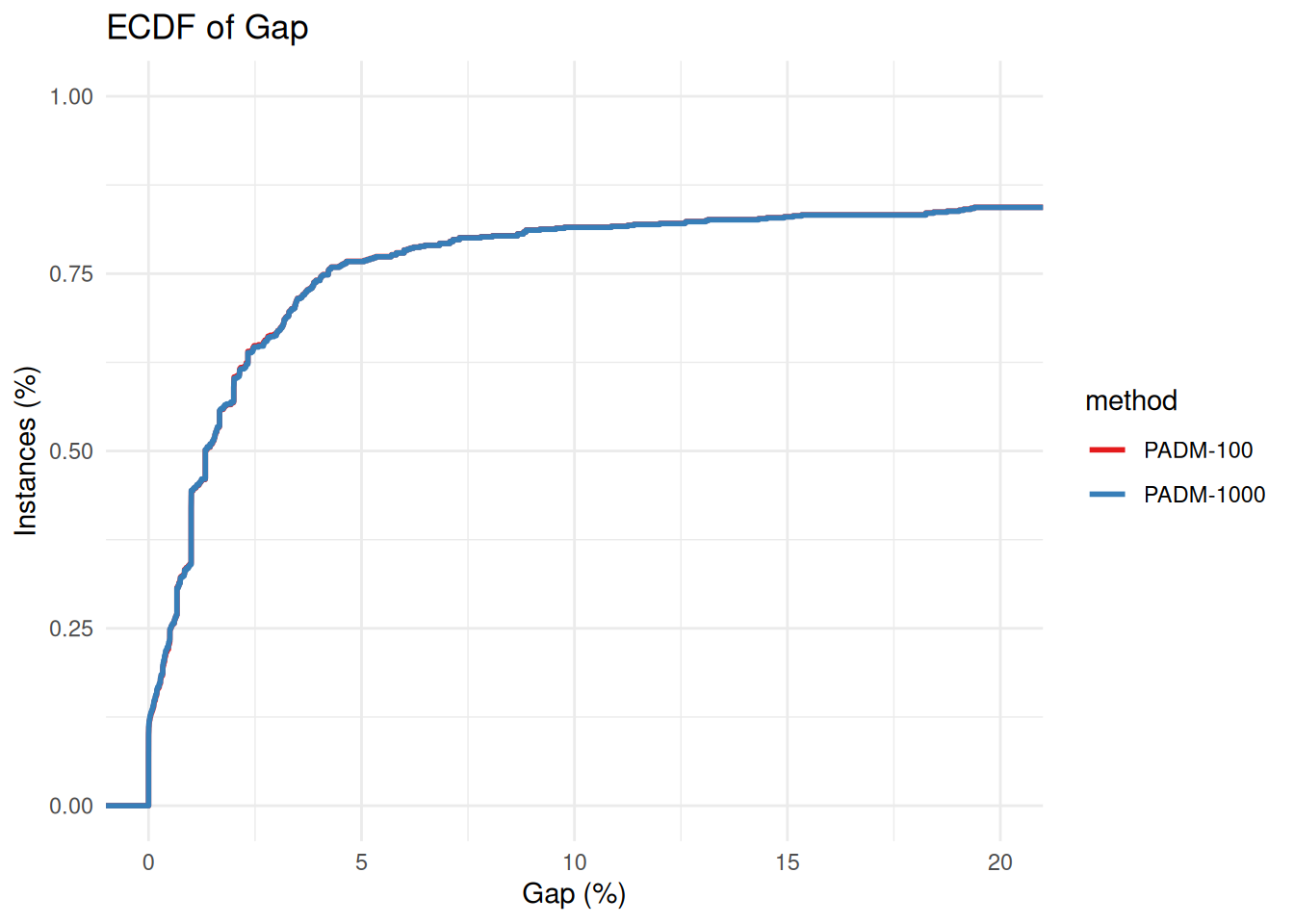
solved_by_both = solved_by_both = all_results %>% filter(method == "PADM-100" | method == "PADM-1000" | method == "MILP-first") %>%
group_by(instance) %>%
filter(all(time < 3600 & time >= 0)) %>%
ungroup()
p = plot_ecdf_of_gap_wrt_best_known(solved_by_both)
p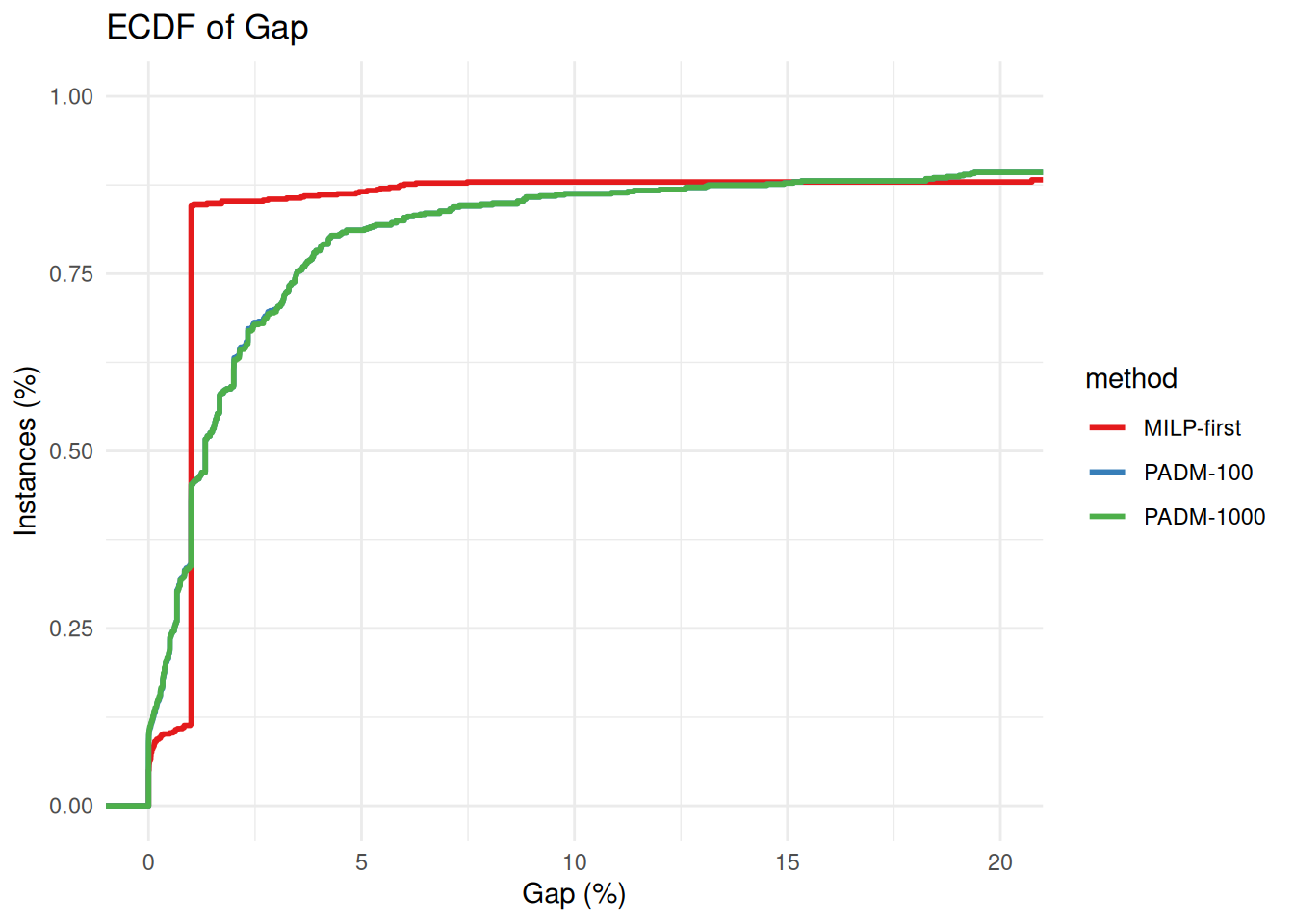
export_data_from_ggplot_ecdf_csv(p, "ecdf_gap_all.csv")Status change in BOBILib
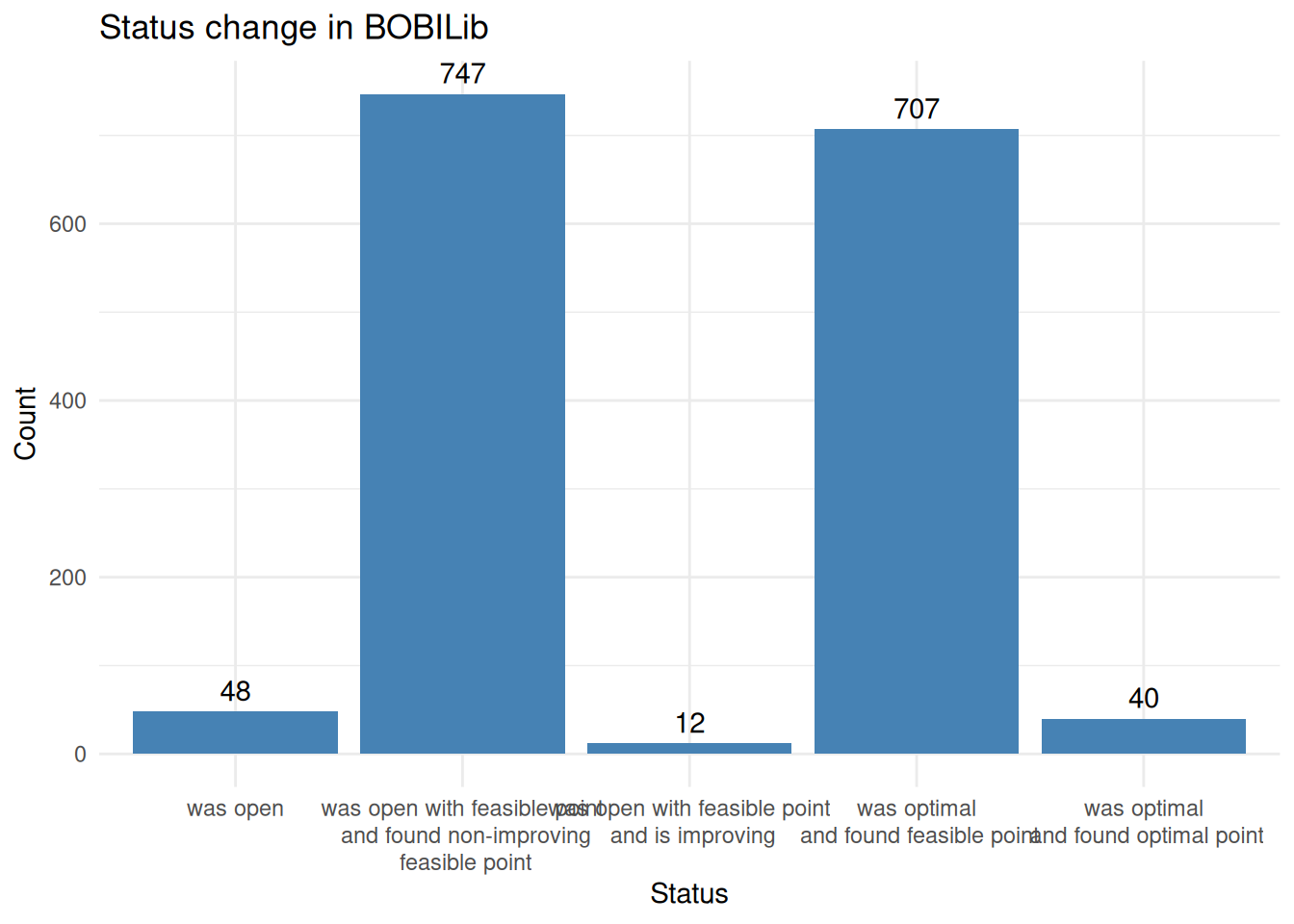
all_results_with_status = all_results %>%
filter(method == "PADM-1000" & time < 3600) %>%
mutate(
new_bobilib_status = ifelse(bobilib_status == "open with feasible point" & best_obj < best_known_obj - 1e-6, "was open with feasible point\n and is improving", bobilib_status),
new_bobilib_status = ifelse(bobilib_status == "optimal" & abs(best_obj - best_known_obj) < 1e-6, "was optimal\n and found optimal point", new_bobilib_status),
new_bobilib_status = ifelse(bobilib_status == "open", "was open", new_bobilib_status),
new_bobilib_status = ifelse(bobilib_status == "optimal" & abs(best_obj - best_known_obj) > 1e-6, "was optimal\n and found feasible point", new_bobilib_status),
new_bobilib_status = ifelse(new_bobilib_status == "open with feasible point" & best_obj >= best_known_obj, "was open with feasible point\n and found non-improving\n feasible point", new_bobilib_status)
)
padm_found_feasible_point = all_results %>%
filter(method == "PADM-1000" & time < 3600)
was_open_and_is_improving = length(padm_found_feasible_point %>% filter(bobilib_status == "open with feasible point" & best_obj < best_known_obj - 1e-6))
was_optimal_and_found_optimal = length(padm_found_feasible_point %>% filter(bobilib_status == "optimal" & abs(best_obj - best_known_obj) < 1e-6))
was_open_and_found_feasible = length(padm_found_feasible_point %>% filter( bobilib_status == "open" ))
was_optimal_and_found_feasible = length(padm_found_feasible_point %>% filter(bobilib_status == "optimal" & abs(best_obj - best_known_obj) > 1e-6))
ggplot(all_results_with_status, aes(x = new_bobilib_status)) +
geom_bar(fill = "steelblue", width = .9) +
geom_text(stat = "count", aes(label = after_stat(count)), vjust = -0.5) +
labs(title = "Status change in BOBILib",
x = "Status",
y = "Count") +
theme_minimal() +
theme(
axis.text.x = element_text(angle = 0, hjust = .5, vjust = 1, lineheight = 1)
)
all_results %>%
filter(method == "PADM-1000" | method == "MILP-first") %>%
filter(time < 3600) %>%
filter( (bobilib_status == "open") ) %>%
select(instance) %>%
unique() %>%
count()## n
## 1 66PADM interdiction
Computational time
all_results_common_to_padm_and_padm_interdiction =
solved_by_both = all_results %>% filter(method == "PADM-1000" | method == "PADM-interdiction-1000") %>%
group_by(instance) %>%
filter(all(time >= 0)) %>%
ungroup()
solved_by_both = solved_by_both = all_results %>% filter(method == "PADM-1000" | method == "PADM-interdiction-1000") %>%
group_by(instance) %>%
filter(all(time < 3600 & time >= 0)) %>%
ungroup()p = plot_ecdf_of_time(all_results_common_to_padm_and_padm_interdiction)
p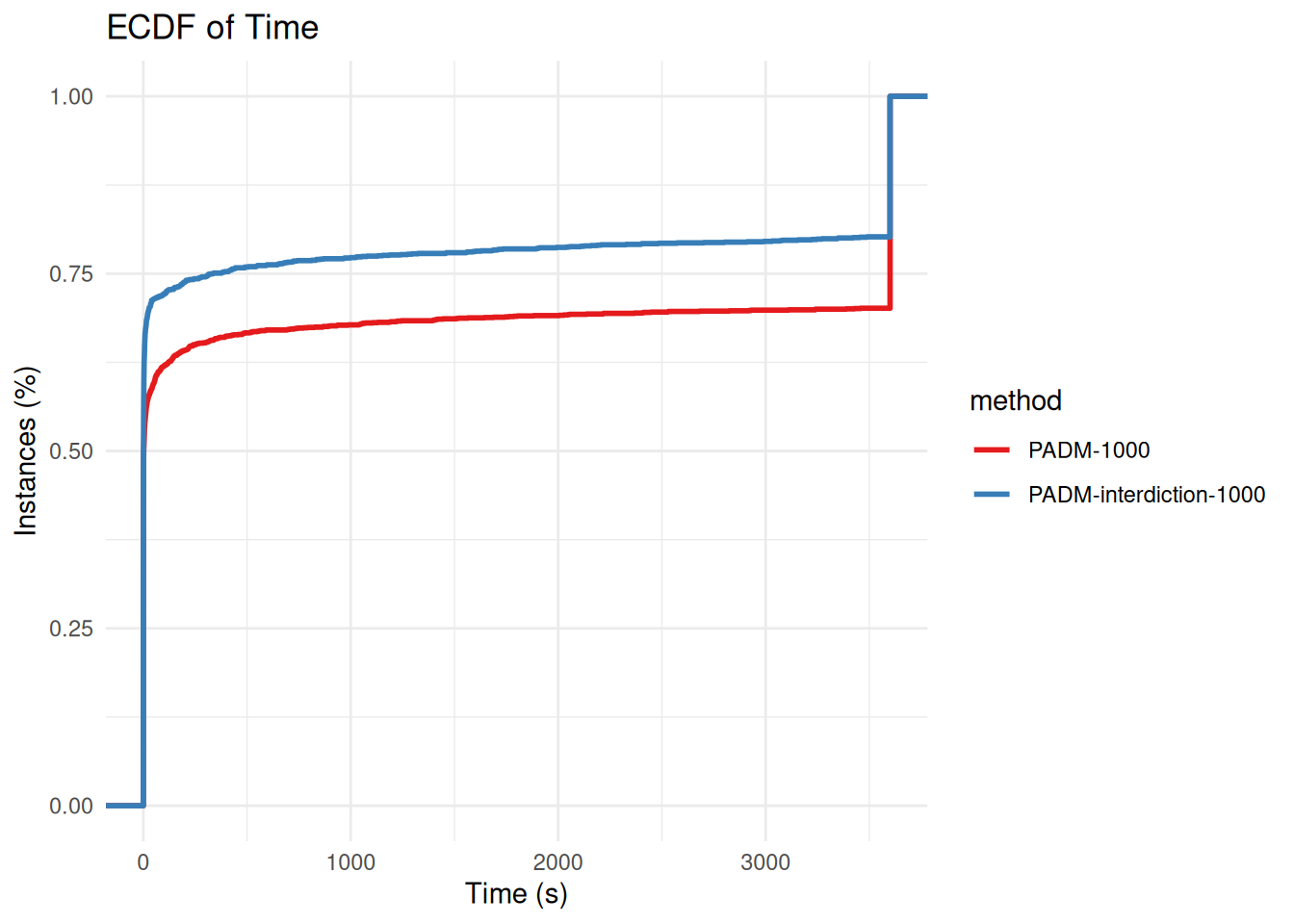
export_data_from_ggplot_ecdf_csv(p, "ecdf_time_interdiction.csv")Optimality gap
plot_ecdf_of_gap_wrt_best_known(all_results %>% filter(method == "PADM-interdiction-1000"))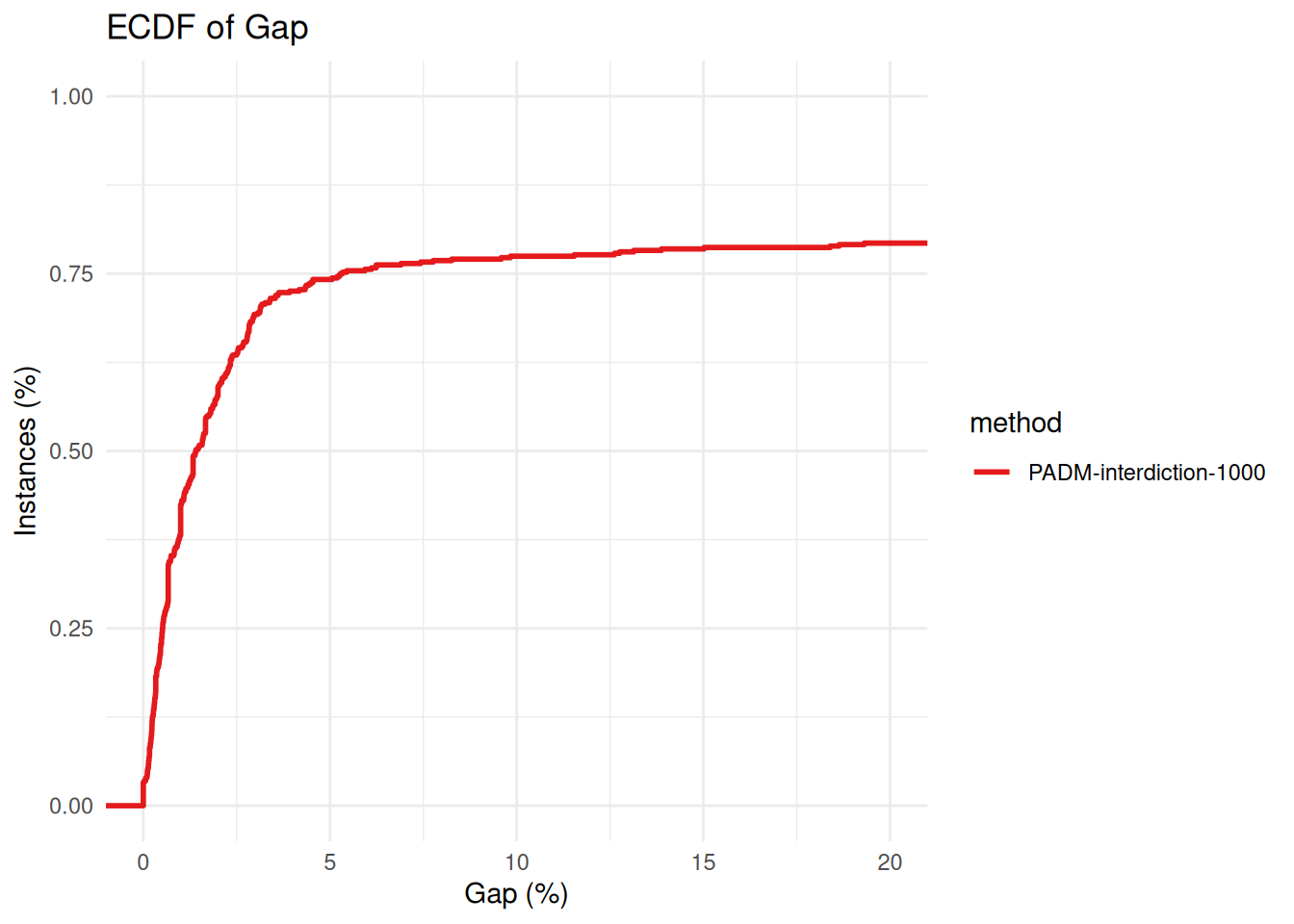
p = plot_ecdf_of_gap_wrt_best_known(solved_by_both)
p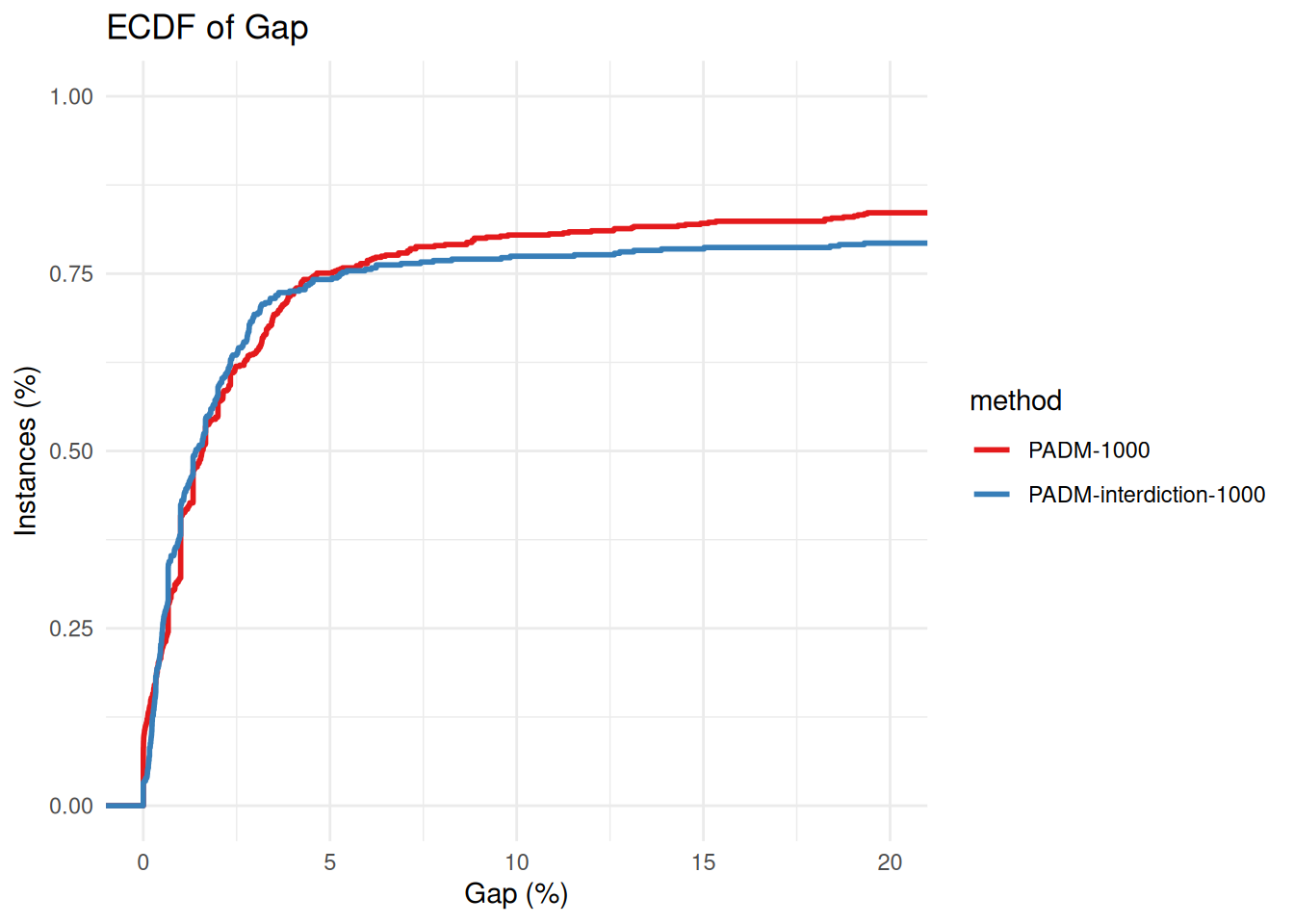
export_data_from_ggplot_ecdf_csv(p, "ecdf_gap_interdiction.csv")#to_check = read.csv("to_check.csv", col.names = c("tag", "instance", "check", "reason"))
#to_check = to_check %>% mutate(instance = basename(instance), instance = gsub("\\.aux$", "", instance))
#all_results %>% filter(method == "MILP-first" & instance %in% to_check$instance & time < 3600)
This document is automatically generated after every
git push action on the public repository
hlefebvr/hlefebvr.github.io using rmarkdown and Github
Actions. This ensures the reproducibility of our data manipulation. The
last compilation was performed on the 20/01/26 18:20:10.
 Henri Lefebvre
Henri Lefebvre